Targeting STK26 and ATG4B: miR-22-3p as a modulator of autophagy and tumor progression in HCC
- PMID: 39608212
- PMCID: PMC11635773
- DOI: 10.1016/j.tranon.2024.102214
Targeting STK26 and ATG4B: miR-22-3p as a modulator of autophagy and tumor progression in HCC
Abstract
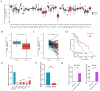
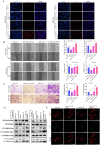
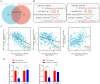
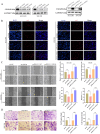
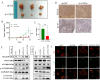
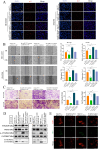
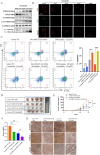
Drug-induced protective autophagy significantly affects the efficacy of anticancer therapies. Enhancing tumor cell sensitivity to treatment by inhibiting autophagy is essential for effective cancer therapy. Our study, analyzing data from The Cancer Genome Atlas (TCGA) public database, HCC cell lines, and liver cancer tissue samples, found that miR-22-3p is expressed at low levels in HCC and is significantly associated with clinicopathological features and patient prognosis. Functional assays and xenograft models demonstrated that miR-22-3p suppresses HCC progression. Moreover, Western blot analysis and the LC3B double reporter (mRFP1-EGFP-LC3B) confirmed that miR-22-3p inhibits autophagy in HCC cells. Further investigation identified Sterile 20-like kinase 26 (STK26) and Autophagy Related 4B Cysteine Peptidase (ATG4B) as targets of miR-22-3p. STK26, which is overexpressed in HCC, promotes malignant characteristics such as proliferation, migration, and invasion. Additionally, STK26 facilitates autophagy in HCC by phosphorylating ATG4B at serine 383. miR-22-3p inhibits autophagy by targeting STK26 and ATG4B, thus preventing the phosphorylation of ATG4B at serine 383. Sorafenib treatment increases the levels and phosphorylation of STK26 and ATG4B, inducing protective autophagy. The combination of miR-22-3p with sorafenib demonstrated enhanced antitumor effects both in vitro and in vivo. In conclusion, our findings suggest that miR-22-3p inhibits HCC progression by regulating the expression of STK26 and ATG4B, potentially through autophagy inhibition, thereby increasing sensitivity to sorafenib treatment. This offers a new therapeutic approach for effective HCC.
Keywords: Autophagy; Hepatocellular carcinoma; Sorafenib; Sterile 20-like kinase 26; miR-22-3p.
Copyright © 2024. Published by Elsevier Inc.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no competing interests.
Figures

Similar articles
-
LncRNA CRNDE Promotes ATG4B-Mediated Autophagy and Alleviates the Sensitivity of Sorafenib in Hepatocellular Carcinoma Cells.Front Cell Dev Biol. 2021 Aug 2;9:687524. doi: 10.3389/fcell.2021.687524. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34409031 Free PMC article.
-
SNGH16 regulates cell autophagy to promote Sorafenib Resistance through suppressing miR-23b-3p via sponging EGR1 in hepatocellular carcinoma.Cancer Med. 2020 Jun;9(12):4324-4338. doi: 10.1002/cam4.3020. Epub 2020 Apr 23. Cancer Med. 2020. PMID: 32324343 Free PMC article.
-
HnRNPU-AS1 inhibits the proliferation, migration and invasion of HCC cells and induces autophagy through miR-556-3p/ miR-580-3p/SOCS6 axis.Cancer Biomark. 2022;34(3):443-457. doi: 10.3233/CBM-210261. Cancer Biomark. 2022. PMID: 35275521
-
PART1 facilitates tumorigenesis and inhibits ferroptosis by regulating the miR-490-3p/SLC7A11 axis in hepatocellular carcinoma.Aging (Albany NY). 2024 Jul 5;16(14):11339-11358. doi: 10.18632/aging.206009. Epub 2024 Jul 5. Aging (Albany NY). 2024. PMID: 39029955 Free PMC article.
-
An ATG4B inhibitor blocks autophagy and sensitizes Sorafenib inhibition activities in HCC tumor cells.Bioorg Med Chem. 2023 Apr 15;84:117262. doi: 10.1016/j.bmc.2023.117262. Epub 2023 Mar 27. Bioorg Med Chem. 2023. PMID: 37018878
Cited by
-
Astragaloside IV inhibits nasopharyngeal carcinoma progression by suppressing the SATB2/Wnt signaling axis.Toxicol Res (Camb). 2025 Apr 2;14(2):tfaf047. doi: 10.1093/toxres/tfaf047. eCollection 2025 Apr. Toxicol Res (Camb). 2025. PMID: 40177383 Free PMC article.
References
-
- Toh M.R., Wong E., Wong S.H., et al. Global epidemiology and genetics of hepatocellular carcinoma. Gastroenterology. 2023;164(5):766–782. - PubMed
-
- Brown Z.J., Tsilimigras D.I., Ruff S.M., et al. Management of hepatocellular carcinoma: a review. JAMA Surg. 2023;158(4):410–420. - PubMed
-
- Yang C., Zhang H., Zhang L., et al. Evolving therapeutic landscape of advanced hepatocellular carcinoma. Nat. Rev. Gastro. Hepat. 2023;20(4):203–222. - PubMed
-
- Ladd A.D., Duarte S., Sahin I., Zarrinpar A. Mechanisms of drug resistance in HCC. Hepatology. 2023 - PubMed
-
- Chen P., Dong Z., Zhu W., et al. Noncanonical regulation of HOIL-1 on cancer stemness and sorafenib resistance identifies pixantrone as a novel therapeutic agent for hepatocellular carcinoma. Hepatology. 2023 - PubMed
LinkOut - more resources
Full Text Sources

