Independent evolution of plant natural products: Formation of benzoxazinoids in Consolida orientalis (Ranunculaceae)
- PMID: 39608711
- PMCID: PMC11742589
- DOI: 10.1016/j.jbc.2024.108019
Independent evolution of plant natural products: Formation of benzoxazinoids in Consolida orientalis (Ranunculaceae)
Abstract
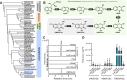
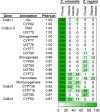
Benzoxazinoids (BXDs) are important defense compounds produced by a number of species from different, evolutionarily unrelated plant families. While BXD biosynthesis has been extensively studied in the grasses (monocots) and core eudicots, the mechanism of BXD synthesis in the basal eudicots is still unclear. We used an integrated metabolomics and transcriptomics approach to elucidate the BXD pathway in Consolida orientalis, a Ranunculaceae species known to produce the BXD DIBOA-Glc. Overexpression of candidate genes in Nicotiana benthamiana identified a flavin-dependent monooxygenase (CoBX2-3) and two cytochrome P450 enzymes (CoBX4 and CoBX5) that catalyze the oxidation steps that transform indole into DIBOA. Co-expression of CoBx2-3, CoBx4, and CoBx5 with the previously described indole synthase gene CoBx1 and the UDP-glucosyltransferase gene CoBx8 in N. benthamiana resulted in the reconstitution of a fully active BXD pathway. The fact that CoBX2-3, CoBX4, and CoBX5 are not phylogenetically related to their counterparts in the grasses and core eudicots suggests independent evolution of benzoxazinoid biosynthesis in these three angiosperm lineages.
Keywords: DIBOA-Glc; benzoxazinoids; biosynthesis; defense compounds; evolution; independent evolution; pathway; plant biochemistry; plant defense; secondary metabolism.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures




References
-
- de Bruijn W.J.C., Gruppen H., Vincken J.-P. Structure and biosynthesis of benzoxazinoids: plant defence metabolites with potential as antimicrobial scaffolds. Phytochemistry. 2018;155:233–243. - PubMed
-
- Niculaes C., Abramov A., Hannemann L., Frey M. Plant protection by benzoxazinoids—recent insights into biosynthesis and function. Agronomy. 2018;8:143.
-
- Frey M., Chomet P., Glawischnig E., Stettner C., Grün S., Winklmair A., et al. Analysis of a chemical plant defense mechanism in grasses. Science. 1997;277:696–699. - PubMed
-
- Baumeler A., Hesse M., Werner C. Benzoxazinoids–cyclic hydroxamic acids, lactams and their corresponding glucosides in the genus Aphelandra (Acanthaceae) Phytochemistry. 2000;53:213–222. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

