GDNF family receptor alpha-like (GFRAL) expression is restricted to the caudal brainstem
- PMID: 39608751
- PMCID: PMC11650321
- DOI: 10.1016/j.molmet.2024.102070
GDNF family receptor alpha-like (GFRAL) expression is restricted to the caudal brainstem
Abstract
Objective: Growth differentiation factor 15 (GDF15) acts on the receptor dimer of GDNF family receptor alpha-like (GFRAL) and Rearranged during transfection (RET). While Gfral-expressing cells are known to be present in the area postrema and nucleus of the solitary tract (AP/NTS) located in the brainstem, the presence of Gfral-expressing cells in other sites within the central nervous system and peripheral tissues is not been fully addressed. Our objective was to thoroughly investigate whether GFRAL is expressed in peripheral tissues and in brain sites different from the brainstem.
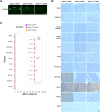
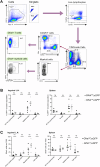
Methods: From Gfral:eGFP mice we collected tissue from 12 different tissues, including brain, and used single molecule in-situ hybridizations to identify cells within those tissues expressing Gfral. We then contrasted the results with human Gfral-expression by analyzing publicly available single-cell RNA sequencing data.
Results: In mice we found readably detectable Gfral mRNA within the AP/NTS but not within other brain sites. Within peripheral tissues, we failed to detect any Gfral-labelled cells in the vast majority of examined tissues and when present, were extremely rare. Single cell sequencing of human tissues confirmed GFRAL-expressing cells are detectable in some sites outside the AP/NTS in an extremely sparse manner. Importantly, across the utilized methodologies, smFISH, genetic Gfral reporter mice and scRNA-Seq, we failed to detect Gfral-labelled cells with all three.
Conclusions: Through highly sensitive and selective technologies we show Gfral expression is overwhelmingly restricted to the brainstem and expect that GDF15 and GFRAL-based therapies in development for cancer cachexia will specifically target AP/NTS cells.
Keywords: Area postrema; GDF15; GFRAL; Nucleus of the solitary tract.
Copyright © 2024 The Author(s). Published by Elsevier GmbH.. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest RJS has received research support from Novo Nordisk, Fractyl, Astra Zeneca, Congruence Therapeutics, Eli Lilly, Bullfrog AI, Glycsend Therapeutics and Amgen. RJS has served as a paid consultant for Novo Nordisk, Eli Lilly, CinRx, Fractyl, Structure Therapeutics, Crinetics and Congruence Therapeutics. RJS has equity in Calibrate, Rewind and Levator Therapeutics. The remaining authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

