Genetic variation at 11q23.1 confers colorectal cancer risk by dysregulation of colonic tuft cell transcriptional activator POU2AF2
- PMID: 39609081
- PMCID: PMC12013567
- DOI: 10.1136/gutjnl-2024-332121
Genetic variation at 11q23.1 confers colorectal cancer risk by dysregulation of colonic tuft cell transcriptional activator POU2AF2
Abstract
Background: Common genetic variation at 11q23.1 is associated with colorectal cancer (CRC) risk, exerting local expression quantitative trait locus (cis-eQTL) effects on POU2AF2, COLCA1 and POU2AF3 genes. However, complex linkage disequilibrium and correlated expression has hindered elucidation of the mechanisms by which genetic variants impart underlying CRC risk.
Objective: Undertake an interdisciplinary approach to understand how variation at 11q23.1 locus imparts CRC risk.
Design: We employ analysis of RNA sequencing, single-cell RNA sequencing, chromatin immunoprecipitation sequencing and single-cell ATAC sequencing data to identify, prioritise and characterise the genes that contribute to CRC risk. We further validate these findings using mouse models and demonstrate parallel effects in human colonic mucosa.
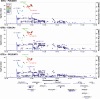
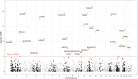
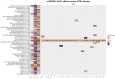
Results: We establish rs3087967 as a prime eQTL variant at 11q23.1, colocalising with CRC risk. Furthermore, rs3087967 influences expression of 21 distant genes, thereby acting as a trans-eQTL hub for a gene-set highly enriched for tuft cell markers. Epigenomic analysis implicates POU2AF2 as controlling the tuft cell-specific trans-genes, through POU2F3-correlated genomic regulation. Immunofluorescence confirms rs3087967 risk genotype (T) to be associated with a tuft cell deficit in the human colon. CRISPR-mediated deletion of the 11q23.1 risk locus genes in the mouse germline exacerbated the ApcMin/+ mouse phenotype on abrogation of Pou2af2 expression specifically.
Conclusion: We demonstrate that genotype at rs3087967 controls a portfolio of genes through misregulation of POU2AF2. POU2AF2 is the primary transcriptional activator of tuft cells with a tumour suppressive role in mouse models. We therefore implicate tuft cells as having a key tumour-protective role in the large bowel epithelium.
Keywords: cancer; cancer susceptibility; colorectal cancer; colorectal cancer genes.
© Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY. Published by BMJ Group.
Conflict of interest statement
Competing interests: None declared.
Figures






References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
