A massively parallel reporter assay library to screen short synthetic promoters in mammalian cells
- PMID: 39609378
- PMCID: PMC11604768
- DOI: 10.1038/s41467-024-54502-9
A massively parallel reporter assay library to screen short synthetic promoters in mammalian cells
Abstract
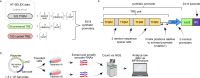
Cellular responses to stimuli underpin discoveries in drug development, synthetic biology, and general life sciences. We introduce a library comprising 6144 synthetic promoters, each shorter than 250 bp, designed as transcriptional readouts of cellular stimulus responses in massively parallel reporter assay format. This library facilitates precise detection and amplification of transcriptional activity from our promoters, enabling the systematic development of tunable reporters with dynamic ranges of 50-100 fold. Our library proved functional in numerous cell lines and responsive to a variety of stimuli, including metabolites, mitogens, toxins, and pharmaceutical agents, generating robust and scalable reporters effective in screening assays, biomarkers, and synthetic circuits attuned to endogenous cellular activities. Particularly valuable in therapeutic development, our library excels in capturing candidate reporters to signals mediated by drug targets, a feature we illustrate across nine diverse G-protein coupled receptors (GPCRs), critical targets in drug development. We detail how this tool isolates and defines discrete signaling pathways associated with specific GPCRs, elucidating their transcriptional signatures. With its ease of implementation, broad utility, publicly available data, and comprehensive documentation, our library will be beneficial in synthetic biology, cellular engineering, ligand exploration, and drug development.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: W.S.O., A.N.G., J.S.B., S.K., and H.C. hold equity in and/or are employed by Octant Inc., a company where similar types of assays are used for its drug discovery efforts. The remaining authors declare no competing interests.
Figures



Update of
-
Discovery and Validation of Context-Dependent Synthetic Mammalian Promoters.bioRxiv [Preprint]. 2023 May 11:2023.05.11.539703. doi: 10.1101/2023.05.11.539703. bioRxiv. 2023. Update in: Nat Commun. 2024 Nov 28;15(1):10353. doi: 10.1038/s41467-024-54502-9. PMID: 37214829 Free PMC article. Updated. Preprint.
References
-
- Dynan, W. S. & Tjian, R. Control of eukaryotic messenger RNA synthesis by sequence-specific DNA-binding proteins. Nature316, 774–778 (1985). - PubMed
-
- Kadonaga, J. T., Carner, K. R., Masiarz, F. R. & Tjian, R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell51, 1079–1090 (1987). - PubMed
-
- Maniatis, T., Goodbourn, S. & Fischer, J. A. Regulation of inducible and tissue-specific gene expression. Science236, 1237–1245 (1987). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

