E3 ubiquitin ligase RNF10 promotes dissociation of stalled ribosomes and responds to ribosomal subunit imbalance
- PMID: 39609413
- PMCID: PMC11604940
- DOI: 10.1038/s41467-024-54411-x
E3 ubiquitin ligase RNF10 promotes dissociation of stalled ribosomes and responds to ribosomal subunit imbalance
Abstract
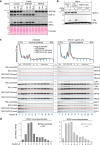
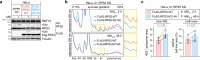
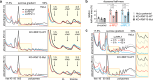
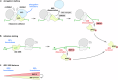
Aberrant translation causes ribosome stalling, which leads to the ubiquitination of ribosomal proteins and induces ribosome-associated quality control. As part of this quality control process, the E3 ubiquitin ligase RNF10 monoubiquitinates ribosomal protein RPS3. Here, we demonstrate that RNF10-mediated RPS3 monoubiquitination antagonizes ribosomal half-mer formation by promoting dissociation of 40S subunits from ribosomes stalled during translation elongation. Interestingly, RNF10 also promotes dissociation of 40S subunits stalled during aberrant translation initiation. Moreover, RNF10 levels are tightly coupled to the amount of 40S subunits. Knockdown of RPS proteins, which abrogates 40S ribosome biogenesis, results in proteasomal degradation of RNF10. Vice versa, knockdown of RPL proteins, which abrogates 60S biogenesis, leads to the accumulation of stalled initiating 40S subunits, increased RNF10 levels, and RPS3 monoubiquitination. As a factor required for the resolution of stalled translation events, RNF10 is part of a fundamental mechanism by which cells respond to imbalances in ribosomal subunit stoichiometry.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Tahmasebi, S., Khoutorsky, A., Mathews, M. B. & Sonenberg, N. Translation deregulation in human disease. Nat. Rev. Mol. Cell Biol.19, 791–807 (2018). - PubMed
-
- Joazeiro, C. A. P. Ribosomal stalling during translation: providing substrates for ribosome-associated protein quality control. Annu. Rev. Cell Dev. Biol.33, 343–368 (2017). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 439669440 - TRR 319/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 464424253 - CRC 1550/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 445549683 - RTG 2727/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 278001972 - TRR 186/Deutsche Forschungsgemeinschaft (German Research Foundation)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

