A DNA methylation signature identified in the buccal mucosa reflecting active tuberculosis is changing during tuberculosis treatment
- PMID: 39609478
- PMCID: PMC11604703
- DOI: 10.1038/s41598-024-80570-4
A DNA methylation signature identified in the buccal mucosa reflecting active tuberculosis is changing during tuberculosis treatment
Abstract
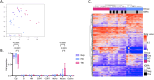
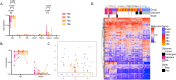
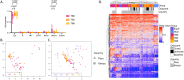
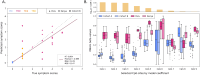
Tuberculosis (TB) poses a significant global health threat, with high mortality rates if left untreated. Current sputum-based TB treatment monitoring methods face numerous challenges, particularly in relation to sample collection and analysis. This pilot study explores the potential of TB status assessment using DNA methylation (DNAm) signatures, which are gaining recognition as diagnostic and predictive tools for various diseases. We collected buccal swab samples from pulmonary TB patients at the commencement of TB treatment (n = 10), and at one, two, and six-month follow-up intervals. We also collected samples from healthy controls (n = 10) and individuals exposed to TB (n = 10). DNAm patterns were mapped using the Illumina Infinium Methylation EPIC 850 K platform. A DNAm profile distinct from controls was discovered in the oral mucosa of TB patients at the start of treatment, and this profile changed throughout the course of TB treatment. These findings were corroborated in a separate validation cohort of TB patients (n = 41), monitored at two and six months into their TB treatment. We developed a machine learning model to predict symptom scores using the identified DNAm TB profile. The model was trained and evaluated on the pilot, validation, and two additional independent cohorts, achieving an R2 of 0.80, Pearson correlation of 0.90, and mean absolute error of 0.13. While validation is needed in larger cohorts, the result opens the possibility of employing DNAm-based diagnostic and prognostic tools for TB in future clinical practice.
Keywords: Biosignatures; Buccal mucosa; DNA methylation; Oral swabs; Treatment monitoring; Tuberculosis.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures
References
-
- Geneva: World Health Organization. Global Tuberculosis Report 2023. January (World Health Organization (WHO), 2023).
-
- World Health Organization. WHO consolidated guidelines on tuberculosis. WHO Press (2022).
-
- World Health Organization. Definitions and reporting framework for tuberculosis - 2013 revision (updated Dec 2014 and Jan 2020) (WHO/ HTM/TB/2013.2). Euro surveillance : bulletin Européen sur les maladies transmissibles = European communicable disease bulletin (2020). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

