Regulation of antimicrobial peptides in Hermetia illucens in response to fungal exposure
- PMID: 39609510
- PMCID: PMC11605112
- DOI: 10.1038/s41598-024-80133-7
Regulation of antimicrobial peptides in Hermetia illucens in response to fungal exposure
Abstract
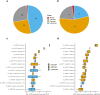
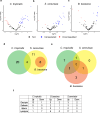
The black soldier fly (Hermetia illucens) is important for antimicrobial peptide (AMP) research due to its exposure to diverse microorganisms. However, the impact of different fungi on AMP abundance in H. illucens remains unexplored. We studied the induction of AMP expression under basal conditions and with three fungi: non-pathogenic Candida tropicalis, Saccharomyces cerevisiae, and pathogenic Beauveria bassiana, using RNA-sequencing and liquid chromatography with tandem mass spectrometry. Under naive conditions, most AMPs belonged to the lysozyme, cecropin, and defensin classes, with defensins most abundant. We demonstrate that dietary supplementation with fungi is sufficient to induce AMP expression in H. illucens. However, exposure to C. tropicalis and B. bassiana also caused downregulation of certain AMPs, suggesting that these fungi may suppress or modulate the host immune response to aid in their survival and colonization. Evidently, S. cerevisiae and B. bassiana trigger similar AMP pathways, whereas C. tropicalis elicits a distinct reaction with upregulation of defensins and cecropins. Lysozymes were upregulated by S. cerevisiae and B. bassiana, but downregulated by C. tropicalis, potentially facilitating fungal survival in the larval gut. Understanding these mechanisms opens possibilities for leveraging AMPs to combat C. tropicalis, which is implicated in human diseases.
Keywords: Beauveria bassiana; Candida tropicalis; Black soldier fly; Defensin; Lysozyme; RNA-seq.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

