Evaluation of large language models for discovery of gene set function
- PMID: 39609565
- PMCID: PMC11725441
- DOI: 10.1038/s41592-024-02525-x
Evaluation of large language models for discovery of gene set function
Abstract
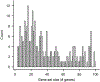
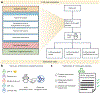
Gene set enrichment is a mainstay of functional genomics, but it relies on gene function databases that are incomplete. Here we evaluate five large language models (LLMs) for their ability to discover the common functions represented by a gene set, supported by molecular rationale and a self-confidence assessment. For curated gene sets from Gene Ontology, GPT-4 suggests functions similar to the curated name in 73% of cases, with higher self-confidence predicting higher similarity. Conversely, random gene sets correctly yield zero confidence in 87% of cases. Other LLMs (GPT-3.5, Gemini Pro, Mixtral Instruct and Llama2 70b) vary in function recovery but are falsely confident for random sets. In gene clusters from omics data, GPT-4 identifies common functions for 45% of cases, fewer than functional enrichment but with higher specificity and gene coverage. Manual review of supporting rationale and citations finds these functions are largely verifiable. These results position LLMs as valuable omics assistants.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests: T.I. is a cofounder and member of the advisory board and has an equity interest in Data4Cure and Serinus Biosciences. T.I. is a consultant for and has an equity interest in Ideaya Biosciences. The terms of these arrangements have been reviewed and approved by the University of California San Diego in accordance with its conflict-of-interest policies. The other authors declare no competing interests.
Figures

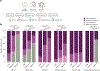


Update of
-
Evaluation of large language models for discovery of gene set function.ArXiv [Preprint]. 2024 Apr 1:arXiv:2309.04019v2. ArXiv. 2024. Update in: Nat Methods. 2025 Jan;22(1):82-91. doi: 10.1038/s41592-024-02525-x. PMID: 37731657 Free PMC article. Updated. Preprint.
-
Evaluation of large language models for discovery of gene set function.Res Sq [Preprint]. 2023 Sep 18:rs.3.rs-3270331. doi: 10.21203/rs.3.rs-3270331/v1. Res Sq. 2023. Update in: Nat Methods. 2025 Jan;22(1):82-91. doi: 10.1038/s41592-024-02525-x. PMID: 37790547 Free PMC article. Updated. Preprint.
References
-
- Beissbarth T & Speed TP GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics 20, 1464–1465 (2004). - PubMed
MeSH terms
Grants and funding
- u01mh115747/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- ot2od023742/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- u24hg12107/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- u24ca269436/Foundation for the National Institutes of Health (Foundation for the National Institutes of Health, Inc.)
- U24 HG012107/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources

