Identification of altered immune cell types and molecular mechanisms in Alzheimer's disease progression by single-cell RNA sequencing
- PMID: 39610716
- PMCID: PMC11602448
- DOI: 10.3389/fnagi.2024.1477327
Identification of altered immune cell types and molecular mechanisms in Alzheimer's disease progression by single-cell RNA sequencing
Abstract
Introduction: Alzheimer's disease (AD) is a progressive neurodegenerative disorder characterized by gradual loss of cognitive function. Understanding the molecular mechanisms is crucial for developing effective therapies.
Methods: Data from single-cell RNA sequencing (scRNA-seq) in the GSE181279 dataset and gene chips in the GSE63060 and GSE63061 datasets were collected and analyzed to identify immune cell types and differentially expressed genes. Cell communication, pseudotime trajectory, enrichment analysis, co- expression network, and short time-series expression miner were analyzed to identify disease-specific molecular and cellular mechanisms.
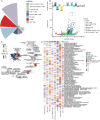
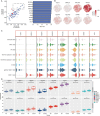
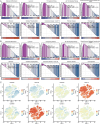
Results: We identified eight cell types (B cells, monocytes, natural killer cells, gamma-delta T cells, CD8+ T cells, Tem/Temra cytotoxic T cells, Tem/Trm cytotoxic T cells, and mucosal-associated invariant T cells) using scRNA-seq. AD samples were enriched in monocytes, CD8+ T cells, Tem/Temra cytotoxic T cells, and Tem/Trm cytotoxic T cells, whereas samples from healthy controls were enriched in natural killer and mucosal-associated invariant T cells. Five co-expression modules that were identified through weighted gene correlation network analysis were enriched in immune- inflammatory pathways. Candidate genes with higher area under the receiver operating characteristic curve values were screened, and the expression trend of Ubiquitin-Fold Modifier Conjugating Enzyme 1 (UFC1) gradually decreased from healthy controls to mild cognitive impairment and then to AD.
Conclusion: Our study suggests that peripheral immune cells may be a potential therapeutic target for AD. Candidate genes, particularly UFC1, may serve as potential biomarkers for progression of AD.
Keywords: Alzheimer’s disease; UFC1; immune cells; monocytes; single-cell RNA sequencing; tlymphocytes.
Copyright © 2024 Lin, Su, Mao, Yang, Huang, Lan, Zeng, Song, Liang, Wei, Zou, Li and Zou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
-
- Davis D. H., Creavin S. T., Noel-Storr A., Quinn T. J., Smailagic N., Hyde C., et al. (2013). Neuropsychological tests for the diagnosis of Alzheimer’s disease dementia and other dementias: A generic protocol for cross-sectional and delayed-verification studies. Cochrane Database Syst. Rev. 2013:CD010460. 10.1002/CD010460 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

