Role of CALCR expression in liver cancer: Implications for the immunotherapy response
- PMID: 39611512
- PMCID: PMC11622006
- DOI: 10.3892/mmr.2024.13406
Role of CALCR expression in liver cancer: Implications for the immunotherapy response
Abstract
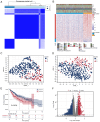
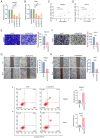
Liver hepatocellular carcinoma (LIHC) is a prevalent and lethal malignancy with a complex molecular landscape. Fibrosis and ferroptosis are implicated in LIHC progression, yet their roles remain to be elucidated. The present study investigated the expression and prognostic significance of calcitonin receptor (CALCR), a gene that intersects the pathways of fibrosis and ferroptosis, across LIHC and other types of cancer. Data were obtained from The Cancer Genome Atlas and the Molecular Signatures Database. LIHC patients were classified into two clusters based on fibrosis‑related gene expression using ConsensusClusterPlus. Single‑sample gene set enrichment analysis was employed to quantify fibrosis and ferroptosis levels. Correlation, survival and nomogram analyses were performed to assess the prognostic value of CALCR. Additionally, single‑cell RNA sequencing data from the Tumor Immune Single Cell Hub 2 (TISCH2) and pan‑cancer analyses of genomic heterogeneity features were incorporated. The present study also identified a putative regulatory role for CALCR in LIHC cell migration, proliferation and apoptosis. CALCR was identified as a significant prognostic marker for LIHC. Patients with high CALCR expression exhibited shortened overall survival (OS) and disease‑specific survival (DSS). Specifically, the hazard ratios (HRs) for OS and DSS were 1.76 [95% confidence interval (CI): 1.23=2.49) and 1.77 (95% CI: 1.13=2.78], respectively, with corresponding P‑values of 0.002 for OS and 0.013 for DSS. Analyses of immune cell infiltration revealed a more complex immune environment in patients with low CALCR expression, suggesting differential responses to immunotherapy. Furthermore, in HepG‑2 and HuH‑7 cells, small interfering (si)‑CALCR increased apoptosis while reducing proliferation and migration compared with si‑negative control. CALCR serves as a significant prognostic biomarker for LIHC, influencing both molecular pathways and the immune landscape. Its expression is associated with improved survival outcomes and distinct genomic features, positioning it as a potential therapeutic target and predictor of immunotherapy efficacy.
Keywords: calcitonin receptor; ferroptosis; fibrosis; genomic heterogeneity; liver hepatocellular carcinoma.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






Similar articles
-
The prognostic and antitumor roles of key genes of ferroptosis in liver hepatocellular cancer and stomach adenocarcinoma.Cancer Biomark. 2024;39(4):335-347. doi: 10.3233/CBM-230114. Cancer Biomark. 2024. PMID: 38393890 Free PMC article.
-
RAC1 serves as a prognostic factor and correlated with immune infiltration in liver hepatocellular carcinoma.J Cancer Res Clin Oncol. 2024 Sep 12;150(9):418. doi: 10.1007/s00432-024-05933-w. J Cancer Res Clin Oncol. 2024. PMID: 39264423 Free PMC article.
-
Analysis of the implication of steroid 5 alpha-reductase 3 on prognosis and immune microenvironment in Liver Hepatocellular Carcinoma.Ann Med. 2024 Dec;56(1):2408463. doi: 10.1080/07853890.2024.2408463. Epub 2024 Sep 28. Ann Med. 2024. PMID: 39340288 Free PMC article.
-
Diagnostic significance and carcinogenic mechanism of pan-cancer gene POU5F1 in liver hepatocellular carcinoma.Cancer Med. 2020 Dec;9(23):8782-8800. doi: 10.1002/cam4.3486. Epub 2020 Sep 26. Cancer Med. 2020. PMID: 32978904 Free PMC article.
-
Role of glutaminyl-peptide cyclo-transferase-like protein (QPCTL) in cancer: From molecular mechanisms to immunotherapy.Gene. 2025 Feb 10;937:149153. doi: 10.1016/j.gene.2024.149153. Epub 2024 Dec 7. Gene. 2025. PMID: 39653089 Review.
Cited by
-
Magnetic resonance imaging bias field correction improves tumor prognostic evaluation after transcatheter arterial chemoembolization for liver cancer.World J Gastrointest Surg. 2025 Apr 27;17(4):104187. doi: 10.4240/wjgs.v17.i4.104187. World J Gastrointest Surg. 2025. PMID: 40291888 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

