T cell receptor-centric perspective to multimodal single-cell data analysis
- PMID: 39612336
- PMCID: PMC11606500
- DOI: 10.1126/sciadv.adr3196
T cell receptor-centric perspective to multimodal single-cell data analysis
Abstract
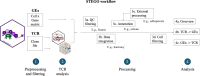
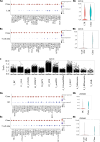
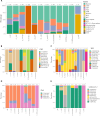
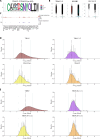
The T cell receptor (TCR), despite its importance, is underutilized in single-cell analysis, with gene expression features solely driving current strategies. Here, we argue for a TCR-first approach, more suited toward T cell repertoires. To this end, we curated a large T cell atlas from 12 prominent human studies, containing in total 500,000 T cells spanning multiple diseases, including melanoma, head and neck cancer, blood cancer, and lung transplantation. Here, we identified severe limitations in cell-type annotation using unsupervised approaches and propose a more robust standard using a semi-supervised method or the TCR arrangement. We showcase the utility of a TCR-first approach through application of the STEGO.R tool for the identification of treatment-related dynamics and previously unknown public T cell clusters with potential antigen-specific properties. Thus, the paradigm shift to a TCR-first can highlight overlooked key T cell features that have the potential for improvements in immunotherapy and diagnostics.
Figures


References
-
- Valkiers S., de Vrij N., Gielis S., Verbandt S., Ogunjimi B., Laukens K., Meysman P., Recent advances in T-cell receptor repertoire analysis: Bridging the gap with multimodal single-cell RNA sequencing. ImmunoInformatics 5, 100009 (2022).
-
- Valkiers S., Van Houcke M., Laukens K., Meysman P., ClusTCR: A python interface for rapid clustering of large sets of CDR3 sequences with unknown antigen specificity. Bioinformatics 37, 4865–4867 (2021). - PubMed
-
- Mayer-Blackwell K., Schattgen S., Cohen-Lavi L., Crawford J. C., Souquette A., Gaevert J. A., Hertz T., Thomas P. G., Bradley P., Fiore-Gartland A., TCR meta-clonotypes for biomarker discovery with tcrdist3 enabled identification of public, HLA-restricted clusters of SARS-CoV-2 TCRs. eLife 10, e68605 (2021). - PMC - PubMed
-
- Glanville J., Huang H., Nau A., Hatton O., Wagar L. E., Rubelt F., Ji X., Han A., Krams S. M., Pettus C., Haas N., Arlehamn C. S. L., Sette A., Boyd S. D., Scriba T. J., Martinez O. M., Davis M. M., Identifying specificity groups in the T cell receptor repertoire. Nature 547, 94–98 (2017). - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

