Identification of HER2-positive breast cancer molecular subtypes with potential clinical implications in the ALTTO clinical trial
- PMID: 39613746
- PMCID: PMC11607438
- DOI: 10.1038/s41467-024-54621-3
Identification of HER2-positive breast cancer molecular subtypes with potential clinical implications in the ALTTO clinical trial
Abstract
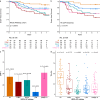
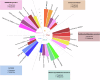
In HER2-positive breast cancer, clinical outcome and sensitivity to HER2-targeted therapies are influenced by both tumor and microenvironment features. However, we are currently unable to depict the molecular heterogeneity of this disease with sufficient granularity. Here, by performing gene expression profiling in HER2-positive breast cancers from patients receiving adjuvant trastuzumab in the ALTTO clinical trial (NCT00490139), we identify and characterize five molecular subtypes associated with the risk of distant recurrence: immune-enriched, proliferative/metabolic-enriched, mesenchymal/stroma-enriched, luminal, and ERBB2-dependent. Additionally, we validate the biological profiles of the subtypes and explore their prognostic/predictive value in external cohorts, namely the NeoALTTO trial (NCT00553358), SCAN-B (NCT02306096), I-SPY2 (NCT01042379), METABRIC and TCGA. Immune-enriched tumors present better survival outcomes, in contrast to mesenchymal/stroma-enriched and proliferative/metabolic-enriched tumors, while luminal and ERBB2-dependent tumors are characterized by low and high rates of pathological complete response, respectively. Of note, these molecular subtypes provide the rationale for treatment approaches leveraging the heterogeneous biology of HER2-positive breast cancer.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare the following competing interests. S.EA.: grants via the affiliation from Novartis for the submitted work and from Roche/Genentech, Astra Zeneca, Pfizer, and BCRF outside the submitted work. S.DC.: consultation fees from Pierre-Fabre, IQVIA, and Medica Scientia Innovation Research (MEDSIR); institutional grant from Fondazione Associazione Italiana Ricerca contro il Cancro (AIRC); and Cancer Can.Heal European EU4 Health Programme 101080009-European Commission. T.U.: honoraria from Astra Zeneca, Novartis Pharma K.K., Eisai Co., Ltd., Chugai Pharmaceutical Co. Ltd; research grant from Eli Lilly Japan K.K. M.I.: employee of Novartis, owner of Novartis shares. M.P.: invited speaker for AstraZeneca, Lilly, MSD, Novartis, Pfizer, Roche-Genentech; consultant for Roche-Genentech; advisory board for Frame Therapeutics, Gilead, Menarini, NBE Therapeutics, Odonate, Roche-Genentech, SeaGen, Seattle Genetics; member of boards of directors, scientific board for Oncolytics; research grants to her Institution from AstraZeneca, Lilly, Gilead; funding to her Institution from Menarini, MSD, Novartis, Pfizer, Radius, Roche-Genentech, Servier, Synthon. L.P.: consulting fees and honoraria for advisory board participation from Pfizer, Astra Zeneca, Merck, Novartis, Bristol-Myers Squibb, Stemline-Menarini, GlaxoSmithKline, Genentech/Roche, Personalis, Daiichi, Natera, Exact Sciences and institutional research funding from Seagen, GlaxoSmithKline, AstraZeneca, Merck, Pfizer and Bristol Myers Squibb. S.L.: research funding to institution from Novartis, Bristol Myers Squibb, MSD, Puma Biotechnology, Eli Lilly, Nektar Therapeutics, Astra Zeneca/Daiichi Sankyo and Seattle Genetics; consultant (not compensated) to Seattle Genetics, Novartis, Bristol Myers Squibb, MSD, AstraZeneca/Daiichi Sankyo, Eli Lilly, Pfizer, Gilead Therapeutics, Nektar Therapeutics, PUMA Biotechnologies, and Roche-Genentech; consultant (paid to institution) to Novartis, GlaxoSmithKline, Roche-Genentech, Astra Zeneca/Daiichi Sankyo, Pfizer, Gilead Therapeutics, Seattle Genetics, MSD, Tallac Therapeutics, Eli Lilly and Bristol Myers Squibb. R.S.: non-financial support from Merck, Case 45, and Bristol Myers Squibb (BMS); research support from Merck, Puma Biotechnology, and Roche; personal fees from Roche, BMS, Astra Zeneca, Daiichi Sankyo and Exact Sciences for advisory boards. G.V.: grants/research supports from Roche/Genentech, Ventana Medical Systems, Dako/Agilent Technologies; honoraria or consultation fees from Ventana, Dako/Agilent, Roche, MSD Oncology, AstraZeneca, Daiichi Sankyo, Pfizer, Gilead. C.S.: advisory board (receipt of honoraria or consultations fees) for Astellas, Cepheid, Vertex, Seattle genetics, Puma, Amgen, Exact Sciences, INC, Merck & Co; invited speaker for Eisai, Prime Oncology, Teva, Foundation Medicine, Exact Sciences; advisory equity from Signatur Biosciences; other support (travel, accommodation expenses) from Roche, Genentech, Pfizer. The remaining authors declare no non-financial or financial competing interests.
Figures



References
-
- Slamon, D. J. et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science235, 177–182 (1987). - PubMed
-
- Cossetti, R. J. D., Tyldesley, S. K., Speers, C. H., Zheng, Y. & Gelmon, K. A. Comparison of breast cancer recurrence and outcome patterns between patients treated from 1986 to 1992 and from 2004 to 2008. J. Clin. Oncol.33, 65–73 (2015). - PubMed
-
- Martin, M. et al. Neratinib after trastuzumab-based adjuvant therapy in HER2-positive breast cancer (ExteNET): 5-year analysis of a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol.18, 1688–1700 (2017). - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

