Multiple myeloma long-term survivors exhibit sustained immune alterations decades after first-line therapy
- PMID: 39613747
- PMCID: PMC11607340
- DOI: 10.1038/s41467-024-54543-0
Multiple myeloma long-term survivors exhibit sustained immune alterations decades after first-line therapy
Abstract
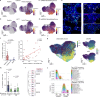
The long-term consequences of cancer and its therapy on the patients' immune system years after cancer-free survival remain poorly understood. Here, we present an in-depth characterization of the bone marrow immune ecosystem of multiple myeloma long-term survivors, from initial diagnosis up to 17 years following a single therapy line and cancer-free survival. Using comparative single-cell analyses combined with molecular, genomic, and functional approaches, we demonstrate that multiple myeloma long-term survivors exhibit pronounced alterations in their bone marrow microenvironment associated with impaired immunity. These immunological alterations were frequently linked to an inflammatory immune circuit fueled by the long-term persistence or resurgence of residual myeloma cells. Notably, even in the complete absence of any detectable residual disease for decades, sustained changes in the immune system were observed, suggesting an irreversible 'immunological scarring' caused by the initial exposure to the cancer and therapy. Collectively, our study provides key insights into the molecular and cellular bone marrow ecosystem of long-term survivors of multiple myeloma, revealing both reversible and irreversible alterations in the immune compartment.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: C.M.-T. received research funding from Bioline RX and Pfizer. M.B. has served as a consultant for Takeda and Novartis; received research funding from Novartis and travel support from Celgene, Amgen, and Janssen. H.G. received funding from the International Myeloma Foundation – Black Swan and the Dietmar-Hopp-Foundation. The other authors declare no competing interests related to this study.
Figures





References
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Medical

