Specific surface-modified iron oxide nanoparticles trigger complement-dependent innate and adaptive antileukaemia immunity
- PMID: 39613769
- PMCID: PMC11607078
- DOI: 10.1038/s41467-024-54810-0
Specific surface-modified iron oxide nanoparticles trigger complement-dependent innate and adaptive antileukaemia immunity
Abstract
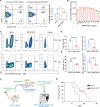
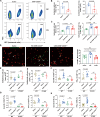
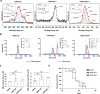
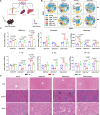
Considerable advances have been achieved in the application of nanomaterials for immunotherapies, yet the precise immune effects induced by protein corona remain elusive. Here, we explore the formation mechanism and immune regulation process of protein corona in acute myeloid leukaemia (AML) mouse models using commercialized iron oxide nanoparticles (IONPs), with different surface modifications, including an FDA-approved variant. Using macrophages depleted or Complement Component 3 (C3) knockout mice, we demonstrate that carboxymethyl dextran-coated IONP (IONP-COOH) reduces leukaemia burden. Mechanistically, IONP-COOH indirectly binds to C3b after activating the complement alternative pathway, subsequently enhancing phagocytosis of macrophages and activating adaptive immunity mediated by complement corona. While aminated dextran-coated IONPs directly absorb C3b and activate the lectin pathway, leading to immune cell exhaustion. Our findings suggest that IONP-COOH may serve as an immune activator for AML treatment, offering a promising approach to developing therapeutic nanomaterials by leveraging surface chemistry to enhance immunotherapy.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Romero, A. et al. Clinical Characteristics of Chilean Adult Patients with Acute Myeloid Leukemia (AML): Analysis within the Framework of the Epidemiological Registry of AML of the Pethema Group. Blood140, 8963–8965 (2022).
-
- Newell, L. F. & Cook, R. J. Advances in acute myeloid leukemia. BMJ375, n2026 (2021). - PubMed
-
- Ci, T. et al. Delivery strategies in treatments of leukemia. Chem. Soc. Rev.51, 2121–2144 (2022). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

