Immune checkpoint landscape of human atherosclerosis and influence of cardiometabolic factors
- PMID: 39613875
- PMCID: PMC11634783
- DOI: 10.1038/s44161-024-00563-4
Immune checkpoint landscape of human atherosclerosis and influence of cardiometabolic factors
Abstract
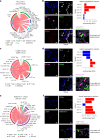
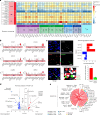
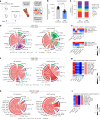
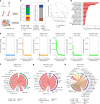
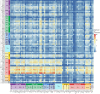
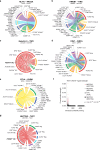
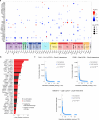
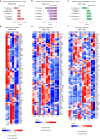
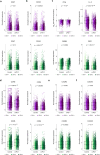
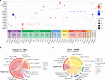
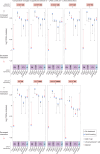
Immune checkpoint inhibitor (ICI) therapies can increase the risk of cardiovascular events in survivors of cancer by worsening atherosclerosis. Here we map the expression of immune checkpoints (ICs) within human carotid and coronary atherosclerotic plaques, revealing a network of immune cell interactions that ICI treatments can unintentionally target in arteries. We identify a population of mature, regulatory CCR7+FSCN1+ dendritic cells, similar to those described in tumors, as a hub of IC-mediated signaling within plaques. Additionally, we show that type 2 diabetes and lipid-lowering therapies alter immune cell interactions through PD-1, CTLA4, LAG3 and other IC targets in clinical development, impacting plaque inflammation. This comprehensive map of the IC interactome in healthy and cardiometabolic disease states provides a framework for understanding the potential adverse and beneficial impacts of approved and investigational ICIs on atherosclerosis, setting the stage for designing ICI strategies that minimize cardiovascular disease risk in cancer survivors.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures







References
-
- FDA approves anti-LAG3 checkpoint. Nat. Biotechnol.10.1038/s41587-022-01331-0 (2022). - PubMed
-
- US FDA. FDA Drug Databasehttps://www.accessdata.fda.gov/scripts/cder/daf/index.cfm (2023).
MeSH terms
Substances
Grants and funding
- R35HL135799/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- R01HL084312/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- P30 CA016087/CA/NCI NIH HHS/United States
- 23POST1029885/American Heart Association (American Heart Association, Inc.)
- R35 HL135799/HL/NHLBI NIH HHS/United States
- R01 HL153712/HL/NHLBI NIH HHS/United States
- 20SFRN35210252/American Heart Association (American Heart Association, Inc.)
- R01HL165258/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- P01 HL151328/HL/NHLBI NIH HHS/United States
- 965509/American Heart Association (American Heart Association, Inc.)
- R01HL153712/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- R01 HL165258/HL/NHLBI NIH HHS/United States
- R01 HL084312/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous
