Meiotic DNA break resection and recombination rely on chromatin remodeler Fun30
- PMID: 39613969
- PMCID: PMC11695836
- DOI: 10.1038/s44318-024-00318-8
Meiotic DNA break resection and recombination rely on chromatin remodeler Fun30
Abstract
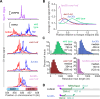
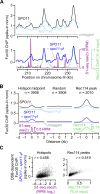
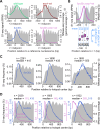
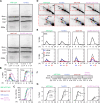
DNA double-strand breaks (DSBs) are nucleolytically processed to generate single-stranded DNA for homologous recombination. In Saccharomyces cerevisiae meiosis, this resection involves nicking by the Mre11-Rad50-Xrs2 complex (MRX), then exonucleolytic digestion by Exo1. Chromatin remodeling at meiotic DSBs is thought necessary for resection, but the remodeling enzyme was unknown. Here we show that the SWI/SNF-like ATPase Fun30 plays a major, nonredundant role in meiotic resection. A fun30 mutation shortened resection tracts almost as severely as an exo1-nd (nuclease-dead) mutation, and resection was further shortened in a fun30 exo1-nd double mutant. Fun30 associates with chromatin in response to DSBs, and the constitutive positioning of nucleosomes governs resection endpoint locations in the absence of Fun30. We infer that Fun30 promotes both the MRX- and Exo1-dependent steps in resection, possibly by removing nucleosomes from broken chromatids. Moreover, the extremely short resection in fun30 exo1-nd double mutants is accompanied by compromised interhomolog recombination bias, leading to defects in recombination and chromosome segregation. Thus, this study also provides insight about the minimal resection lengths needed for robust recombination.
Keywords: Exo1; Fun30; Meiosis; Recombination; Resection.
© 2024. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures






Update of
-
Meiotic DNA break resection and recombination rely on chromatin remodeler Fun30.bioRxiv [Preprint]. 2024 Apr 18:2024.04.17.589955. doi: 10.1101/2024.04.17.589955. bioRxiv. 2024. Update in: EMBO J. 2025 Jan;44(1):200-224. doi: 10.1038/s44318-024-00318-8. PMID: 38659928 Free PMC article. Updated. Preprint.
References
-
- Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P (1997) An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386:414–417 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

