Genomic and phenotypic characterization of ST2012 clinical Klebsiella quasipneumoniae subsp. similipneumoniae harboring blaNDM-1 in China
- PMID: 39614148
- PMCID: PMC11606014
- DOI: 10.1186/s12866-024-03637-2
Genomic and phenotypic characterization of ST2012 clinical Klebsiella quasipneumoniae subsp. similipneumoniae harboring blaNDM-1 in China
Abstract
Background: Klebsiella pneumoniae has emerged as a significant pathogen that causes community and hospital infections due to its high resistance rate and transmissibility. Klebsiella pneumoniae complex is classified into three phylogroups: Klebsiella pneumoniae (KpI), Klebsiella quasipneumoniae (KpII), and Klebsiella variicola (KpIII) in classical taxonomy, but KpII and KpIII are infrequently isolated clinically. Although pathogenic KpII has been reported worldwide, the understanding of this pathogen remains limited.
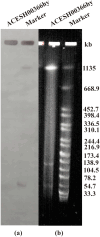
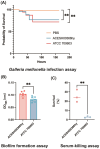
Methods: Whole-genome sequencing (WGS) of K. quasipneumoniae subsp. similipneumoniae ACESH00366hy was performed. Plasmid characterization was demonstrated using S1-PFGE, Southern blotting, and conjugation assays. Antimicrobial susceptibility testing was performed and interpreted according to CLSI, EUCAST, and FDA standards. The virulence of the strain was assessed using the Galleria mellonella infection assay, serum-killing assay, and biofilm formation assay. A phylogenetic tree was constructed to explore its biological evolution.
Results: The K. quasipneumoniae subsp. similipneumoniae isolate ACESH00366hy, belonging to ST2012 and KL139, contains several resistance genes including blaNDM-1, blaSHV-12, blaOKP-B-2, and oqxAB, as well as various virulence genes including iroE, iutA, mrkABCDFHIJ, entABCDEFS, and fepABCDG. The blaNDM-1 and blaSHV-12 genes were present on the 53,624 bp IncX3 plasmid. Virulence assays showed that the virulence of ACESH00366hy was greater than that of the low-toxicity strain ATCC 700,603. Phylogenetic analysis demonstrated the emergence of ST2012 KpII-B in Asia and revealed the spread of K. quasipneumoniae subsp. similipneumoniae in humans, animals, and the environment.
Conclusion: This study highlights the emergence of clinical ST2012 KpII strains harbouring blaNDM-1 via the IncX3 plasmid in China. We evaluated its resistance and virulence characteristics and performed phylogenetic analysis, thereby enhancing our understanding of its resistance and pathogenicity and contributing to the clinical surveillance of K. quasipneumoniae.
Keywords: Klebsiella pneumoniae; Klebsiella quasipneumoniae; bla NDM−1; KpII.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: The study was approved by the Ethics Committee of the First Affiliated Hospital of Zhejiang University School of Medicine (reference number: 2021-IIT-631). This was a retrospective study and patient consent was not required. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Characterization of a multidrug-resistant hypovirulent ST1859-KL35 klebsiella quasipneumoniae subsp. similipneumoniae strain co-harboring tmexCD2-toprJ2 and blaKPC-2.J Glob Antimicrob Resist. 2025 May;42:253-261. doi: 10.1016/j.jgar.2025.03.009. Epub 2025 Mar 18. J Glob Antimicrob Resist. 2025. PMID: 40113085
-
Whole genome sequencing reveals antibiotic resistance pattern and virulence factors in Klebsiella quasipneumoniae subsp. Similipneumoniae from Hospital wastewater in South-West, Nigeria.Microb Pathog. 2024 Dec;197:107040. doi: 10.1016/j.micpath.2024.107040. Epub 2024 Oct 19. Microb Pathog. 2024. PMID: 39427715
-
Description of Klebsiella quasipneumoniae sp. nov., isolated from human infections, with two subspecies, Klebsiella quasipneumoniae subsp. quasipneumoniae subsp. nov. and Klebsiella quasipneumoniae subsp. similipneumoniae subsp. nov., and demonstration that Klebsiella singaporensis is a junior heterotypic synonym of Klebsiella variicola.Int J Syst Evol Microbiol. 2014 Sep;64(Pt 9):3146-3152. doi: 10.1099/ijs.0.062737-0. Epub 2014 Jun 23. Int J Syst Evol Microbiol. 2014. PMID: 24958762
-
Genomic and molecular characterization of a ceftazidime-avibactam resistant Klebsiella pneumoniae strain isolated from a Chinese tertiary hospital.BMC Microbiol. 2025 Apr 8;25(1):199. doi: 10.1186/s12866-025-03929-1. BMC Microbiol. 2025. PMID: 40200176 Free PMC article.
-
From soil to surface water: exploring Klebsiella 's clonal lineages and antibiotic resistance odyssey in environmental health.BMC Microbiol. 2025 Feb 27;25(1):97. doi: 10.1186/s12866-025-03798-8. BMC Microbiol. 2025. PMID: 40012032 Free PMC article. Review.
Cited by
-
Genomic dataset of multidrug-resistant Klebsiella quasipneumoniae from Indonesia.BMC Res Notes. 2025 May 15;18(1):218. doi: 10.1186/s13104-025-07269-1. BMC Res Notes. 2025. PMID: 40375280 Free PMC article.
References
-
- Rosenblueth M, Martinez L, Silva J, Martinez-Romero E. Klebsiella variicola, a novel species with clinical and plant-associated isolates. Syst Appl Microbiol. 2004;27(1):27–35. - PubMed
-
- Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A. 2015;112(27):E3574–3581. - PMC - PubMed
-
- Fonseca EL, Ramos ND, Andrade BG, Morais LL, Marin MF, Vicente AC. A one-step multiplex PCR to identify Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae in the clinical routine. Diagn Microbiol Infect Dis. 2017;87(4):315–7. - PubMed
-
- Imai K, Ishibashi N, Kodana M, Tarumoto N, Sakai J, Kawamura T, Takeuchi S, Taji Y, Ebihara Y, Ikebuchi K, et al. Clinical characteristics in blood stream infections caused by Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae: a comparative study, Japan, 2014–2017. BMC Infect Dis. 2019;19(1):946. - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
- 2020YFE0204300/National Key Research and Development Program of China
- 82072314/National Natural Science Foundation of China
- LHDMZ22H190002/Zhejiang Provincial Natural Science Foundation of China
- SYS202202/Shandong Provincial Laboratory Project
- JNL-2022011B/Research Project of Jinan Microecological Biomedicine Shandong Laboratory
LinkOut - more resources
Full Text Sources
Medical

