The central role of creatine and polyamines in fetal growth restriction
- PMID: 39614665
- PMCID: PMC11607630
- DOI: 10.1096/fj.202401946R
The central role of creatine and polyamines in fetal growth restriction
Abstract
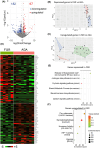
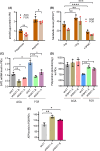
Placental insufficiency often correlates with fetal growth restriction (FGR), a condition that has both short- and long-term effects on the health of the newborn. In our study, we analyzed placental tissue from infants with FGR and from infants classified as small for gestational age (SGA) or appropriate for gestational age (AGA), performing comprehensive analyses that included transcriptomics and metabolomics. By examining villus tissue biopsies and 3D trophoblast organoids, we identified significant metabolic changes in placentas associated with FGR. These changes include adaptations to reduced oxygen levels and modifications in arginine metabolism, particularly within the polyamine and creatine phosphate synthesis pathways. Specifically, we found that placentas with FGR utilize arginine to produce phosphocreatine, a crucial energy reservoir for ATP production that is essential for maintaining trophoblast function. In addition, we found polyamine insufficiency in FGR placentas due to increased SAT1 expression. SAT1 facilitates the acetylation and subsequent elimination of spermine and spermidine from trophoblasts, resulting in a deficit of polyamines that cannot be compensated by arginine or polyamine supplementation alone, unless SAT1 expression is suppressed. Our study contributes significantly to the understanding of metabolic adaptations associated with placental dysfunction and provides valuable insights into potential therapeutic opportunities for the future.
Keywords: creatine metabolism; fetal growth restriction; metabolome; placenta biopsies; placenta organoids; polyamine metabolism; spermidine/spermidine N1‐acetyltransferase 1; transcriptome.
© 2024 The Author(s). The FASEB Journal published by Wiley Periodicals LLC on behalf of Federation of American Societies for Experimental Biology.
Figures





References
-
- Society for Maternal‐Fetal Medicine (SMFM). Electronic address: pubs@smfm.org , Miller R, Timor‐Tritsch IE, Gyamfi‐Bannerman C. Society for Maternal‐Fetal Medicine Consult Series #52: diagnosis and management of fetal growth restriction: (replaces clinical guideline number 3, April 2012). Am J Obstet Gynecol. 2020;223(4):B2‐B17. - PubMed
-
- Fetal growth restriction: ACOG practice bulletin, number 227. Obstet Gynecol. 2021;137(2):e16‐e28. - PubMed
-
- Lees CC, Stampalija T, Baschat AA, et al. ISUOG practice guidelines: diagnosis and management of small‐for‐gestational‐age fetus and fetal growth restriction. Ultrasound Obstet Gynecol. 2020;56(2):298‐312. - PubMed
-
- Defining normal and abnormal fetal growth: promises and challenges – ScienceDirect [Internet]. Accessed May 14, 2024. https://www.sciencedirect.com/science/article/pii/S0002937809021152.
-
- Gordijn SJ, Beune IM, Thilaganathan B, et al. Consensus definition of fetal growth restriction: a Delphi procedure. Ultrasound Obstet Gynecol. 2016;48(3):333‐339. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

