Apoptotic signaling by TNFR1 is inhibited by the α2-6 sialylation, but not α2-3 sialylation, of the TNFR1 N-glycans
- PMID: 39615678
- PMCID: PMC11732462
- DOI: 10.1016/j.jbc.2024.108043
Apoptotic signaling by TNFR1 is inhibited by the α2-6 sialylation, but not α2-3 sialylation, of the TNFR1 N-glycans
Abstract
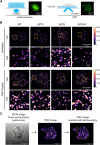
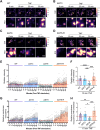
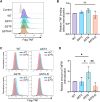
The TNF-TNFR1 signaling pathway plays a pivotal role in regulating the balance between cell survival and cell death. Upon binding to TNF, plasma membrane-localized TNFR1 initiates survival signaling, whereas TNFR1 internalization promotes caspase-mediated apoptosis. We previously reported that the α2-6 sialylation of TNFR1 by the tumor-associated sialyltransferase ST6GAL1 diverts signaling toward survival by inhibiting TNFR1 internalization. In the current investigation, we interrogated the mechanisms underlying sialylation-dependent regulation of TNFR1 and uncovered a novel role for α2-6 sialylation, but not α2-3 sialylation, in mediating apoptosis-resistance. Our studies utilized HEK293 cells with deletion of sialyltransferases that modify N-glycans with either α2-3-linked sialic acids (ST3GAL3/4/6) or α2-6-linked sialic acids (ST6GAL1/2). Additionally, ST6GAL1 was re-expressed in cells with ST6GAL1/2 deletion to restore α2-6 sialylation. Using total internal reflection fluorescence (TIRF) microscopy and BS3 cross-linking, we determined that, under basal conditions, cells expressing TNFR1 devoid of α2-6 sialylation displayed enhanced TNFR1 oligomerization, an event that poises cells for activation by TNF. Moreover, upon stimulation with TNF, greater internalization of TNFR1 was observed via time-lapse TIRF and flow cytometry, and this correlated with increased caspase-dependent apoptosis. These effects were reversed by ST6GAL1 re-expression. Conversely, eliminating α2-3 sialylation did not significantly alter TNFR1 clustering, internalization or apoptosis. We also evaluated the Fas receptor, given its structural similarity to TNFR1. As with TNFR1, α2-6 sialylation had a selective effect in protecting cells against Fas-mediated apoptosis. These results collectively suggest that ST6GAL1 may serve a unique function in shielding cancer cells from apoptotic stimuli within the tumor microenvironment.
Keywords: Fas; ST6GAL1; TNFR1; apoptosis; cancer; death receptors; glycosylation; sialic acid.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

