Multiomic Network Analysis Identifies Dysregulated Neurobiological Pathways in Opioid Addiction
- PMID: 39615775
- PMCID: PMC12119972
- DOI: 10.1016/j.biopsych.2024.11.013
Multiomic Network Analysis Identifies Dysregulated Neurobiological Pathways in Opioid Addiction
Abstract
Background: Opioid addiction is a worldwide public health crisis. In the United States, for example, opioids cause more drug overdose deaths than any other substance. However, opioid addiction treatments have limited efficacy, meaning that additional treatments are needed.
Methods: To help address this problem, we used network-based machine learning techniques to integrate results from genome-wide association studies of opioid use disorder and problematic prescription opioid misuse with transcriptomic, proteomic, and epigenetic data from the dorsolateral prefrontal cortex of people who died of opioid overdose and control individuals.
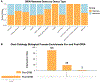
Results: We identified 211 highly interrelated genes identified by genome-wide association studies or dysregulation in the dorsolateral prefrontal cortex of people who died of opioid overdose that implicated the Akt, BDNF (brain-derived neurotrophic factor), and ERK (extracellular signal-regulated kinase) pathways, identifying 414 drugs targeting 48 of these opioid addiction-associated genes. Some of the identified drugs are approved to treat other substance use disorders or depression.
Conclusions: Our synthesis of multiomics using a systems biology approach revealed key gene targets that could contribute to drug repurposing, genetics-informed addiction treatment, and future discovery.
Keywords: Addiction; Bioinformatics; Multiomic; Networks; Opioids; Systems biology.
Copyright © 2025. Published by Elsevier Inc.
Figures
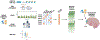


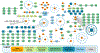
References
-
- UN Office on Drugs and Crime (2021): World Drug Report: Drug Market Trends: Cannabis, Opioids. Available at: https://www.unodc.org/unodc/en/data-and-analysis/wdr2021.html. Accessed December 21, 2022.
-
- Substance Abuse and Mental Health Services Administration: Key substance use and mental health indicators in the United States: Results from the 2020 National Survey on Drug Use and Health. Available at: https://www.samhsa.gov/data/sites/default/files/reports/rpt35319/2020NSD.... Accessed December 21, 2022.
-
- Spencer M, Miniño A, Warner M (2022): Drug overdose deaths in the United States, 2001–2021. National Center for Health Statistics. Available at: https://www.cdc.gov/nchs/data/databriefs/db457.pdf. Accessed December 21, 2022.
-
- Ahmad FB, Cisewski JA, Rossen LM, Sutton P (2023): Provisional drug overdose death counts. National Center for Health Statistics. Available at: https://www.cdc.gov/nchs/nvss/vsrr/drug-overdose-data.htm. Accessed December 21, 2022.
-
- United States Department of Health and Human Services: HHS by the numbers: Combating the opioids crisis. Available at: https://www.hhs.gov/sites/default/files/hhs-by-the-numbers-opioidssept-2.... Accessed December 21, 2022.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

