SenMayo transcriptomic senescence panel highlights glial cells in the ageing mouse and human retina
- PMID: 39616175
- PMCID: PMC11608257
- DOI: 10.1038/s41514-024-00187-9
SenMayo transcriptomic senescence panel highlights glial cells in the ageing mouse and human retina
Abstract
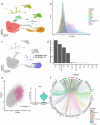
There is a growing need to better characterise senescent cells in the CNS and retina. The recently published SenMayo gene panel was developed to identify transcriptomic signatures of senescence across multiple organ systems, but the retina was not included. While other approaches have identified senescent signatures in the retina, these have largely focused on experimental models in young animals. We therefore conducted a detailed single-cell RNA-seq analysis to identify senescent cell populations in the retina of different aged mice and compared these with five comprehensive human and mouse retina and brain transcriptome datasets. Transcriptomic signatures of senescence were most apparent in mouse and human retinal glial cells, with IL4, 13 and 10 and the AP1 pathway being the most prominent markers involved. Similar levels of transcriptional senescence were observed in the retinal glia of young and old mice, whereas the human retina showed significantly increased enrichment scores with advancing age.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval: All breeding and experimental procedures were undertaken in accordance with the Association for Research for Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Research and were approved by the SingHealth Institutional Animal Care and Use Committee (2019/SHS/1534).
Figures






References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

