Characterization of radiations-induced genomic structural variations in Arabidopsis thaliana
- PMID: 39616610
- PMCID: PMC11712536
- DOI: 10.1111/tpj.17180
Characterization of radiations-induced genomic structural variations in Arabidopsis thaliana
Abstract
DNA, is assaulted by endogenous and exogenous agents that lead to the formation of damage. In order to maintain genome integrity DNA repair pathways must be efficiently activated to prevent mutations and deleterious chromosomal rearrangements. Conversely, genome rearrangement is also necessary to allow genetic diversity and evolution. The antagonist interaction between maintenance of genome integrity and rearrangements determines genome shape and organization. Therefore, it is of great interest to understand how the whole linear genome structure behaves upon formation and repair of DNA damage. For this, we used long reads sequencing technology to identify and to characterize genomic structural variations (SV) of wild-type Arabidopsis thaliana somatic cells exposed either to UV-B, to UV-C or to protons irradiations. We found that genomic regions located in heterochromatin are more prone to form SVs than those located in euchromatin, highlighting that genome stability differs along the chromosome. This holds true in Arabidopsis plants deficient for the expression of master regulators of the DNA damage response (DDR), ATM (Ataxia-telangiectasia-mutated) and ATR (Ataxia-telangiectasia-mutated and Rad3-related), suggesting that independent and alternative surveillance processes exist to maintain integrity in genic regions. Finally, the analysis of the radiations-induced deleted regions allowed determining that exposure to UV-B, UV-C and protons induced the microhomology-mediated end joining mechanism (MMEJ) and that both ATM and ATR repress this repair pathway.
Keywords: DSB repair; genome flexibility; genome stability; genotoxic stress; ionizing radiations; non‐ionizing radiations; structural variants.
© 2024 The Author(s). The Plant Journal published by Society for Experimental Biology and John Wiley & Sons Ltd.
Conflict of interest statement
We declare no competing financial interests.
Figures


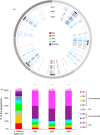
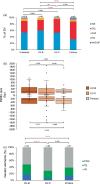

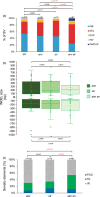
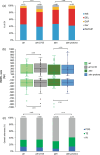

Similar articles
-
An insight into the mechanism of DNA damage response in plants- role of SUPPRESSOR OF GAMMA RESPONSE 1: An overview.Mutat Res. 2020 Jan-Apr;819-820:111689. doi: 10.1016/j.mrfmmm.2020.111689. Epub 2020 Jan 23. Mutat Res. 2020. PMID: 32004947 Review.
-
An insight into understanding the coupling between homologous recombination mediated DNA repair and chromatin remodeling mechanisms in plant genome: an update.Cell Cycle. 2021 Sep;20(18):1760-1784. doi: 10.1080/15384101.2021.1966584. Epub 2021 Aug 26. Cell Cycle. 2021. PMID: 34437813 Free PMC article. Review.
-
Quantitative phosphoproteomics of the ataxia telangiectasia-mutated (ATM) and ataxia telangiectasia-mutated and rad3-related (ATR) dependent DNA damage response in Arabidopsis thaliana.Mol Cell Proteomics. 2015 Mar;14(3):556-71. doi: 10.1074/mcp.M114.040352. Epub 2015 Jan 5. Mol Cell Proteomics. 2015. PMID: 25561503 Free PMC article.
-
Arabidopsis ATM and ATR kinases prevent propagation of genome damage caused by telomere dysfunction.Plant Cell. 2011 Dec;23(12):4254-65. doi: 10.1105/tpc.111.092387. Epub 2011 Dec 9. Plant Cell. 2011. PMID: 22158468 Free PMC article.
-
DNA damage checkpoint kinase ATM regulates germination and maintains genome stability in seeds.Proc Natl Acad Sci U S A. 2016 Aug 23;113(34):9647-52. doi: 10.1073/pnas.1608829113. Epub 2016 Aug 8. Proc Natl Acad Sci U S A. 2016. PMID: 27503884 Free PMC article.
References
-
- Ata, H. , Ekstrom, T.L. , Martínez‐Gálvez, G. , Mann, C.M. , Dvornikov, A.V. , Schaefbauer, K.J. et al. (2018) Robust activation of microhomology‐mediated end joining for precision gene editing applications. PLoS Genetics, 9, e1007652. Available from: 10.1371/journal.pgen.1007652 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

