Spatial transcriptomics deciphers the immunosuppressive microenvironment in colorectal cancer with tumour thrombus
- PMID: 39617739
- PMCID: PMC11608866
- DOI: 10.1002/ctm2.70112
Spatial transcriptomics deciphers the immunosuppressive microenvironment in colorectal cancer with tumour thrombus
Conflict of interest statement
The authors declare no conflict of interest.
Figures

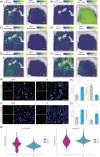
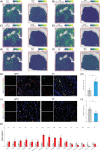
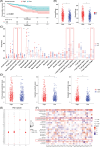
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
