BRAF inhibitors enhance erythropoiesis and treat anemia through paradoxical activation of MAPK signaling
- PMID: 39617757
- PMCID: PMC11609275
- DOI: 10.1038/s41392-024-02033-6
BRAF inhibitors enhance erythropoiesis and treat anemia through paradoxical activation of MAPK signaling
Abstract
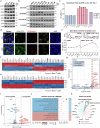
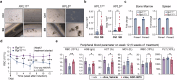
Erythropoiesis is a crucial process in hematopoiesis, yet it remains highly susceptible to disruption by various diseases, which significantly contribute to the global challenges of anemia and blood shortages. Current treatments like erythropoietin (EPO) or glucocorticoids often fall short, especially for hereditary anemias such as Diamond-Blackfan anemia (DBA). To uncover new erythropoiesis-stimulating agents, we devised a screening system using primary human hematopoietic stem and progenitor cells (HSPCs). We discovered that BRAF inhibitors (BRAFi), commonly used to treat BRAFV600E melanoma, can unexpectedly and effectively promote progenitor cell proliferation by temporarily delaying erythroid differentiation. Notably, these inhibitors exhibited pronounced efficacy even under cytokine-restricted conditions and in patient samples of DBA. Mechanistically, although these BRAFi inhibit the MAPK cascade in BRAFV600E mutant cells, they paradoxically act as amplifiers in wild-type BRAF cells, potently enhancing the cascade. Furthermore, we found that while the oncogenic BRAFV600E mutation disrupts hematopoiesis and erythropoiesis through AP-1 hyperactivation, BRAFi minimally impact HSPC self-renewal and differentiation. In vivo studies have shown that BRAFi can enhance human hematopoiesis and erythropoiesis in severe immunodeficient mouse models and alleviate anemia in the Rpl11 haploinsufficiency DBA model, as well as other relevant anemia models. This discovery underscores the role of the MAPK pathway in hematopoiesis and positions BRAFi as a promising therapeutic option for improving hematopoietic reconstitution and treating anemias, including DBA.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Schematic figures were generated with BioRender ( https://app.biorender.com/ ).
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

