Cofactor recycling strategies for secondary metabolite production in cell-free protein expression systems
- PMID: 39618802
- PMCID: PMC11604874
- DOI: 10.1007/s12551-024-01234-1
Cofactor recycling strategies for secondary metabolite production in cell-free protein expression systems
Abstract
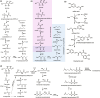
Cell-free protein synthesis (CFPS) has emerged as an attractive platform for biotechnology and synthetic biology due to its numerous advantages to cell-based technologies for specific applications. CFPS can be faster, less sensitive to metabolite toxicity, and amenable to systems that are not easily genetically manipulated. Due to these advantages, a promising application of CFPS is to characterize biosynthetic gene clusters, particularly those harbored within the genomes of microorganisms that generate secondary metabolites, otherwise known as natural products. In the postgenomic era, genome sequencing has revealed an incredible wealth of metabolic diversity. However, far more of these pathways are termed "cryptic," i.e., unable to be produced under standard laboratory conditions than have been characterized. A major barrier to characterizing these cryptic natural products using CFPS is that many of these pathways require utilization of complex cofactors, many of which to date are not recycled efficiently or in an economically viable fashion. In this perspective, we outline strategies to regenerate cofactors relevant to secondary metabolite production in CFPS. This includes adenosine 5'-triphosphate, coenzyme A, redox cofactors (iron-sulfur clusters, nicotinamide adenine dinucleotide phosphate, flavin adenine dinucleotide), all of which play a crucial role in important biosynthetic enzymes. Such advances in cofactor recycling enable continuous production of complex metabolites in CFPS and expand the utility of this emergent platform.
Keywords: Cell Free Protein Synthesis; Cofactor recycling strategies; Secondary metabolite production.
© The Author(s) 2024.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures




References
-
- Alissandratos A, Caron K, Loan TD, Hennessy JE, Easton CJ (2016) ATP recycling with cell lysate for enzyme-catalyzed chemical synthesis, protein expression and PCR. ACS Chem Biol 11(12):3289–3293. 10.1021/acschembio.6b00838 - PubMed
-
- Bartzoka N, Loan TD, Onagi H, Alissandratos A (2022) A simple recombinant E. coli cell lysate-based biocatalyst for ATP-dependent multi-step reactions. Methods Mol Biol 2487:297–315. 10.1007/978-1-0716-2269-8_18 - PubMed
-
- Bao J, Ryu DD (2007) Total biosynthesis of deoxynucleoside triphosphates using deoxynucleoside monophosphate kinases for PCR application. Biotechnol Bioeng 98(1):1–11. 10.1002/bit.21498 - PubMed
-
- Begley TP, Kinsland C, Strauss E (2001) The biosynthesis of coenzyme A in bacteria. Vitam Horm 61:157–171. 10.1016/s0083-6729(01)61005-7 - PubMed
-
- Beinert H, Holm RH, Münck E (1997) Iron-sulfur clusters: nature’s modular multipurpose structures. Science 277(5326):653–659. 10.1126/science.277.5326.653 - PubMed
Publication types
LinkOut - more resources
Full Text Sources

