JUN and ATF3 in Gout: Ferroptosis-related potential diagnostic biomarkers
- PMID: 39619595
- PMCID: PMC11605342
- DOI: 10.1016/j.heliyon.2024.e39957
JUN and ATF3 in Gout: Ferroptosis-related potential diagnostic biomarkers
Abstract
Objective: Gout is a prevalent form of chronic inflammatory arthritis, and its etiology remains incompletely understood. Ferroptosis is a form of cell death that relies on iron. As of now, the relationship between ferroptosis and gout is not entirely clear. Hence, the primary objective of this study is to employ bioinformatics methods for the analysis and identification of potential genes associated with ferroptosis in the context of gout.
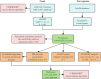
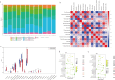
Methods: Utilizing both bioinformatics analysis and machine learning algorithms to systematically identify biomarkers for gout. The gout-related dataset (GSE160170) was acquired from the Gene Expression Omnibus (GEO) database. Ferroptosis-related genes were extracted from the FerrDb database. subsequently, we identified DEGs associated with ferroptosis in the context of gout. Following that, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses on the DEGs. Subsequently, SVM-RFE analysis and the LASSO regression model were employed for biomarker screening. Additionally, CIBERSORT software was utilized to assess the composition of twenty-two immune cells in gout, and correlation analyses between hub genes and immune cells were conducted.
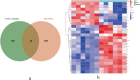
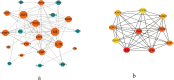
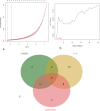
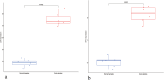
Results: This study screened a total of twenty-five DEGs related to Ferroptosis in healthy population and gout patient. The KEGG analysis indicates that these DEGs are predominantly enriched in: the AGE-RAGE signaling pathway, nod like receptor signaling pathway, MAPK signaling pathway, IL-17 signaling pathway, etc. The intersection of the top 10 genes identified through PPI network, SVM-RFE analysis, and LASSO regression model resulted in two hub genes, namely JUN and ATF3. Analysis of immunocyte infiltration revealed that JUN exhibited associations with various immune cells, including NK cells resting, Monocytes, Mast cells resting, etc. ATF3, on the other hand, showed associations with immune cells Mast cells resting and Eosinophels.
Conclusions: The outcomes of our study pinpointed JUN and ATF3, genes associated with ferroptosis, as promising biomarkers for both diagnosing and treating gout, providing additional evidence to support the important role of ferroptosis in gout and providing potential therapeutic methods for clinical targeted ferroptosis prevention and treatment of gout.
Keywords: Bioinformatics; Biomarkers; Ferroptosis; Gout; Machine learning.
© 2024 The Authors.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this paper.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

