Origin and evolution of West Nile virus lineage 1 in Italy
- PMID: 39620707
- PMCID: PMC11626449
- DOI: 10.1017/S0950268824001420
Origin and evolution of West Nile virus lineage 1 in Italy
Abstract
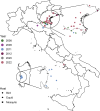
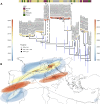
West Nile virus (WNV) is a mosquito-borne pathogen that can infect humans, equids, and many bird species, posing a threat to their health. It consists of eight lineages, with Lineage 1 (L1) and Lineage 2 (L2) being the most prevalent and pathogenic. Italy is one of the hardest-hit European nations, with 330 neurological cases and 37 fatalities in humans in the 2021-2022 season, in which the L1 re-emerged after several years of low circulation. We assembled a database comprising all publicly available WNV genomes, along with 31 new Italian strains of WNV L1 sequenced in this study, to trace their evolutionary history using phylodynamics and phylogeography. Our analysis suggests that WNV L1 may have initially entered Italy from Northern Africa around 1985 and indicates a connection between European and Western Mediterranean countries, with two distinct strains circulating within Italy. Furthermore, we identified new genetic mutations that are typical of the Italian strains and that can be tested in future studies to assess their pathogenicity. Our research clarifies the dynamics of WNV L1 in Italy, provides a comprehensive dataset of genome sequences for future reference, and underscores the critical need for continuous and coordinated surveillance efforts between Europe and Africa.
Keywords: Europe; Italy; West Nile virus; lineage 1; mosquito; phylogeography.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Rizzoli A, et al. (2015) The challenge of West Nile virus in Europe: knowledge gaps and research priorities. Euro Surveillance 20, 21135. - PubMed
-
- Kramer LD, et al. (2007). West Nile virus. The Lancet Neurology 6, 171–181. - PubMed
-
- Smithburn KC, et al. (1940) A neurotropic virus isolated from the blood of a native of Uganda. American Journal of Tropical Medicine 20, 471–472.
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

