HO-1 impairs the efficacy of radiotherapy by redistributing cGAS and STING in tumors
- PMID: 39621310
- PMCID: PMC11601901
- DOI: 10.1172/JCI181044
HO-1 impairs the efficacy of radiotherapy by redistributing cGAS and STING in tumors
Abstract
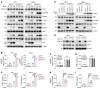
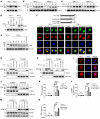
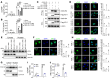
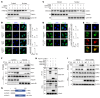
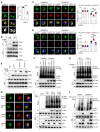
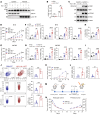
Type I IFNs (IFN-Is) induced by radiotherapy (RT) are critical for its efficacy, while the mechanism by which tumor cells inhibit IFN-I production remains largely unsolved. By an unbiased CRISPR screen, we identified hemeoxygenase 1 (HO-1) as an RT-related regulator of IFN-I production. Mechanistically, the ER-anchored, full-length HO-1 disrupted stimulator of IFN genes (STING) polymerization and subsequent coat protein complex II-mediated (COPII-mediated) ER-Golgi transportation, leading to hampered activation of downstream signaling. This process was exacerbated by the upregulation of HO-1 expression under RT. Importantly, RT also induced HO-1 cleavage. Cleaved HO-1 underwent nuclear translocation, interacted with cyclic GMP-AMP synthase (cGAS), and inhibited its nuclear export upon irradiation, leading to suppressed 2'3'-cyclic GMP-AMP (cGAMP) production. Furthermore, we revealed that HO-1 inhibitors could enhance local and distant tumor control of RT in vivo. Clinically, higher HO-1 expression was associated with a poorer prognosis and earlier tumor relapse after RT in multiple types of patient tumors. Collectively, through comprehensive inhibition of the cGAS/STING pathway, HO-1 strongly inhibited RT-induced IFN-I production, and targeting HO-1 was shown to be a promising RT-sensitizing therapeutic strategy.
Keywords: Cellular immune response; Immunology; Oncology; Radiation therapy.
Figures




Comment in
- Taking the STING out of radiotherapy: STING checkpoints mediate radiation resistance doi: 10.1172/JCI186547
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

