Dysregulated Treg repair responses lead to chronic rejection after heart transplantation
- PMID: 39621314
- PMCID: PMC11601918
- DOI: 10.1172/JCI173593
Dysregulated Treg repair responses lead to chronic rejection after heart transplantation
Abstract
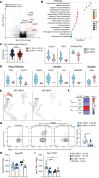
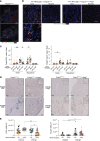
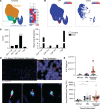
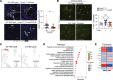
Chronic rejection (CR) after organ transplantation is alloimmune injury manifested by graft vascular remodeling and fibrosis that is resistant to immunosuppression. Single-cell RNA-Seq analysis of MHC class II-mismatched (MHCII-mismatched) heart transplants developing chronic rejection identified graft IL-33 as a stimulator of tissue repair pathways in infiltrating macrophages and Tregs. Using IL-33-deficient donor mice, we show that graft fibroblast-derived IL-33 potently induced amphiregulin (Areg) expression by recipient Tregs. The assessment of clinical samples also confirmed increased expression of Areg by intragraft Tregs also during rejection. Areg is an EGF secreted by multiple immune cells to shape immunomodulation and tissue repair. In particular, Areg is proposed to play a major role in Treg-mediated muscle, epithelium, and nerve repair. Assessment of recipient mice with Treg-specific deletion of Areg surprisingly uncovered that Treg secretion of Areg contributed to CR. Specifically, heart transplants from recipients with Areg-deficient Tregs showed less fibrosis, vasculopathy, and vessel-associated fibrotic niches populated by recipient T cells. Mechanistically, we show that Treg-secreted Areg functioned to increase fibroblast proliferation. In total, these studies identify how a dysregulated repair response involving interactions between IL-33+ fibroblasts in the allograft and recipient Tregs contributed to the progression of CR.
Keywords: Cellular immune response; Fibrosis; Immunology; Organ transplantation; Transplantation.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

