Single nucleotide polymorphisms are associated with strain-specific virulence differences among clinical isolates of Cryptococcus neoformans
- PMID: 39622806
- PMCID: PMC11612297
- DOI: 10.1038/s41467-024-54729-6
Single nucleotide polymorphisms are associated with strain-specific virulence differences among clinical isolates of Cryptococcus neoformans
Erratum in
-
Author Correction: Single nucleotide polymorphisms are associated with strain-specific virulence differences among clinical isolates of Cryptococcus neoformans.Nat Commun. 2025 Mar 13;16(1):2510. doi: 10.1038/s41467-025-57919-y. Nat Commun. 2025. PMID: 40082434 Free PMC article. No abstract available.
Abstract
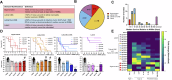
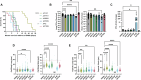
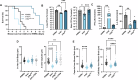
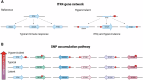
Studies across various pathogens highlight the importance of pathogen genetic differences in disease manifestation. In the human fungal pathogen Cryptococcus neoformans, sequence type (ST) associates with patient outcome. We performed a meta-analysis of four genomic studies and identified overlapping gene regions associated with virulence, suggesting the importance of these gene regions in cryptococcal disease in diverse clinical isolates. We explored the relationship between virulence and strain genetic differences using the cryptococcosis mouse model and a closely related library of ST93 clinical isolates. We identified four in vivo virulence phenotypes: hypervirulence, typical virulence with CNS disease, typical virulence with non-CNS disease, and latent disease. Hypervirulent isolates were clade specific and associated with an interferon gamma (IFNγ) dominated immune response. Using a genome wide association study (GWAS), we identified nine genes with polymorphisms associated with IFNγ production, including the inositol sensor ITR4. The itr4Δ mutant recapitulated the hypervirulence phenotype and ITR4 affects expression of two IFNγ associated genes. Finally, we showed that IFNγ production is associated with SNPs that downregulate ITR4 and with SNP accumulation in other IFNγ associated genes. These data highlight the complex role of pathogen genetics in virulence and identify genes associated with hypervirulence and IFNγ in Cryptococcus neoformans.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
- F31AI148047/Division of Intramural Research, National Institute of Allergy and Infectious Diseases (Division of Intramural Research of the NIAID)
- R01AI123315/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R21NS108715/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01AI134636/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01AI155647/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01 NS118538/NS/NINDS NIH HHS/United States
- R01 AI176922/AI/NIAID NIH HHS/United States
- R01 AI123315/AI/NIAID NIH HHS/United States
- F31 AI148047/AI/NIAID NIH HHS/United States
- R01 AI155647/AI/NIAID NIH HHS/United States
- R01NS118538/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01 AI134636/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources

