Mechanisms of life cycle simplification in African trypanosomes
- PMID: 39622840
- PMCID: PMC11612274
- DOI: 10.1038/s41467-024-54555-w
Mechanisms of life cycle simplification in African trypanosomes
Abstract
African trypanosomes are important parasites in sub-Saharan Africa that undergo a quorum-sensing dependent development to morphologically 'stumpy forms' in mammalian hosts to favour transmission by tsetse flies. However, some trypanosome clades have simplified their lifecycle by escaping dependence on tsetse allowing an expanded geographic range, with direct transmission between hosts achieved via blood-feeding biting flies and vampire bats (Trypanosoma brucei evansi, causing 'surra') or through sexual transmission (Trypanosoma brucei equiperdum, causing 'dourine'). Concomitantly, stumpy formation is reduced and the isolates are described as monomorphic, with infections spread widely in Africa, Asia, South America and parts of Europe. Here, using genomic analysis of distinct field isolates, we identify molecular changes that accompany the loss of the stumpy formation in monomorphic clades. Using CRISPR-mediated allelic replacement, mutations in two exemplar genes (Tb927.2.4020; Tb927.5.2580) are confirmed to reduce stumpy formation whereas another (Tb927.11.3400) is implicated in altered motility. Using laboratory selection we identify downregulation of RNA regulators as important in the initial development of monomorphism. This identifies a trajectory of events that simplify the life cycle in emergent and established monomorphic trypanosomes, with impact on disease spread, vector control strategies, geographical range and virulence.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

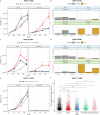
 ; T. b. brucei vs add-back,
; T. b. brucei vs add-back,  ). * (p < 0.05); ** (p < 0.01; *** (p < 0.001); **** (p < 0.0001). For the growth and IFA analysis, four flasks were grown, three biological replicates for growth analysis and one to screen PAD1 and K/N counts at 48 h. f The motility of the FAZ41 replacement cell lines was also quantified for cells expressing the T. b. brucei (n = 1758), T. b. equiperdum type BoTat (n = 2618), T. b. equiperdum type OVI (n = 2003), T. b. equiperdum type BoTat:add-back (n = 867) and T. b. equiperdum type OVI:add-back (n = 3192) each compared between three biological replicates using a Wilcoxon two-sample test and the significance is indicated (* (p < 0.05); ** (p < 0.01; *** (p < 0.001); **** (p < 0.0001)). Source data are provided as a Source Data file.
). * (p < 0.05); ** (p < 0.01; *** (p < 0.001); **** (p < 0.0001). For the growth and IFA analysis, four flasks were grown, three biological replicates for growth analysis and one to screen PAD1 and K/N counts at 48 h. f The motility of the FAZ41 replacement cell lines was also quantified for cells expressing the T. b. brucei (n = 1758), T. b. equiperdum type BoTat (n = 2618), T. b. equiperdum type OVI (n = 2003), T. b. equiperdum type BoTat:add-back (n = 867) and T. b. equiperdum type OVI:add-back (n = 3192) each compared between three biological replicates using a Wilcoxon two-sample test and the significance is indicated (* (p < 0.05); ** (p < 0.01; *** (p < 0.001); **** (p < 0.0001)). Source data are provided as a Source Data file.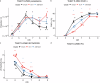
 ; brucei vs add-back,
; brucei vs add-back,  ). * (p < 0.05); ** (p < 0.01; *** (p < 0.001); **** (p < 0.0001). d Representative DAPI, PAD1 and merged images of each cell line at 6 DPI. Scale bar = 30 µm. Source data are provided as a Source Data file.
). * (p < 0.05); ** (p < 0.01; *** (p < 0.001); **** (p < 0.0001). d Representative DAPI, PAD1 and merged images of each cell line at 6 DPI. Scale bar = 30 µm. Source data are provided as a Source Data file.

References
-
- Bruce, D., Hamerton, A. E., Bateman, H. R. & Mackie, F. P. Trypanosome diseases of domestic animals in Uganda II.—Trypanosoma brucei (Plimmer and Bradford). Proc. R. Soc. Lond. B Biol. Sci.83, 1–14 (1910). - DOI
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources

