A novel small molecule Enpp1 inhibitor improves tumor control following radiation therapy by targeting stromal Enpp1 expression
- PMID: 39622844
- PMCID: PMC11612208
- DOI: 10.1038/s41598-024-80677-8
A novel small molecule Enpp1 inhibitor improves tumor control following radiation therapy by targeting stromal Enpp1 expression
Abstract
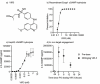
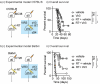
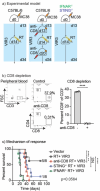
The uniqueness in each person's cancer cells and variation in immune infiltrates means that each tumor represents a unique problem, but therapeutic targets can be found among their shared features. Radiation therapy alters the interaction between the cancer cells and the stroma through release of innate adjuvants. The extranuclear DNA that can result from radiation damage of cells can result in production of the second messenger cyclic guanosine monophosphate-adenosine monophosphate (cGAMP) by cyclic GMP-AMP synthase (cGAS). In turn, cGAMP can activate the innate sensor stimulator of interferon genes (STING), resulting in innate immune activation. Ectonucleotide pyrophosphatase/phosphodiesterase 1 (Enpp1) is a phosphodiesterase that can be expressed by cancer cells that can degrade cGAMP, thus can decrease or block STING activation following radiation therapy, impairing the innate immunity that is critical to support adaptive immune control of tumors. We observed that many human and murine cancer cells lack Enpp1 expression, but that Enpp1 is expressed in cells of the tumor stroma where it limits tumor control by radiation therapy. We demonstrate in preclinical models the efficacy of a novel Enpp1 inhibitor and show that this inhibitor improves tumor control by radiation even where the cancer cells lack Enpp1. This mechanism requires STING and type I interferon (IFN) receptor expression by non-cancer cells and is dependent on CD8 T cells as a final effector mechanism of tumor control. This suggests that Enpp1 inhibition may be an effective partner for radiation therapy regardless of whether cancer cells express Enpp1. This broadens the potential patient base for whom Enpp1 inhibitors can be applied to improve innate immune responses following radiation therapy.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The work was funded in part by a sponsored research agreement with VIR Biotechnology Inc. In addition, the research was funded in part by NCI R01CA182311, NCI R01CA244142, R01CA208644, R21CA277250, and by the Providence Foundation. MJG receives research funding from Bristol Myers Squibb. MRC receives consulting fees from Roche. Authors listed as employees of VIR Biotechnology Inc. and may own stock or other interests in VIR Biotechnology. Authors have no additional conflicts to declare.
Figures




References
-
- Yang, H. et al. RG7204 (PLX4032), a selective BRAFV600E inhibitor, displays potent antitumor activity in preclinical melanoma models. Cancer Res.70, 5518–5527. 10.1158/0008-5472.Can-10-0646 (2010). - PubMed
-
- Garcia, J. et al. Bevacizumab (Avastin(R)) in cancer treatment: a review of 15 years of clinical experience and future outlook. Cancer Treat. Rev.86, 102017. 10.1016/j.ctrv.2020.102017 (2020). - PubMed
-
- Pages, F. et al. International validation of the consensus immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet391, 2128–2139. 10.1016/S0140-6736(18)30789-X (2018). - PubMed
-
- Galon, J. et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science313, 1960–1964. 10.1126/science.1129139 (2006). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

