Regulatory T cells crosstalk with tumor cells and endothelium through lymphotoxin signaling
- PMID: 39622857
- PMCID: PMC11612289
- DOI: 10.1038/s41467-024-54874-y
Regulatory T cells crosstalk with tumor cells and endothelium through lymphotoxin signaling
Abstract
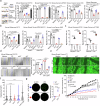
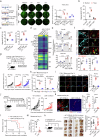
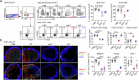
Regulatory T cells (Tregs) with multifaceted functions suppress anti-tumor immunity by signaling surrounding cells. Here we report Tregs use the surface lymphotoxin (LT)α1β2 to preferentially stimulate LT beta receptor (LTβR) nonclassical NFκB signaling on both tumor cells and lymphatic endothelial cells (LECs) to accelerate tumor growth and metastasis. Selectively targeting LTβR nonclassical NFκB pathway inhibits tumor growth and migration in vitro. Leveraging in vivo Treg LTα1β2 interactions with LTβR on tumor cells and LECs, transfer of wild type but not LTα-/- Tregs promotes B16F10 melanoma growth and tumor cell-derived chemokines in LTβR-/- mice; and increases SOX18 and FLRT2 in lymphatic vessels of LTβR-/- melanoma. Blocking the nonclassical pathway suppresses tumor growth and lymphatic metastasis by reducing chemokine production, restricting Treg recruitment to tumors, and retaining intratumoral IFNγ+ CD8 T cells. Our data reveals that Treg LTα1β2 promotes LTβR nonclassical NFκB signaling in tumor cells and LECs providing a rational strategy to prevent Treg promoted tumor growth and metastasis.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: J.S.B., W.P., and X.Y. are inventors on a patent related to LTβR antagonists entitled “Inhibitors of LTβR-NFκB signaling pathways for treating inflammation and cancer” (Patent number: 11590202). The remaining authors declare no competing interests.
Figures





References
-
- Lukashev, M. et al. Targeting the lymphotoxin-beta receptor with agonist antibodies as a potential cancer therapy. Cancer Res.66, 9617–9624 (2006). - PubMed
-
- Gommerman, J. L. & Browning, J. L. Lymphotoxin/light, lymphoid microenvironments and autoimmune disease. Nat. Rev. Immunol.3, 642–655 (2003). - PubMed
-
- Wolf, M. J., Seleznik, G. M., Zeller, N. & Heikenwalder, M. The unexpected role of lymphotoxin beta receptor signaling in carcinogenesis: from lymphoid tissue formation to liver and prostate cancer development. Oncogene29, 5006–5018 (2010). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

