Proteomic analysis of plasma and duodenal tissue in celiac disease patients reveals potential noninvasive diagnostic biomarkers
- PMID: 39622892
- PMCID: PMC11612483
- DOI: 10.1038/s41598-024-80391-5
Proteomic analysis of plasma and duodenal tissue in celiac disease patients reveals potential noninvasive diagnostic biomarkers
Abstract
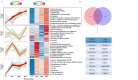
The pathogenesis of celiac disease (CeD) remains incompletely understood. Traditional diagnostic techniques for CeD include serological testing and endoscopic examination; however, they have limitations. Therefore, there is a need to identify novel noninvasive biomarkers for CeD diagnosis. We analyzed duodenal and plasma samples from CeD patients by four-dimensional data-dependent acquisition (4D-DIA) proteomics. Differentially expressed proteins (DEPs) were identified for functional analysis and to propose blood biomarkers associated with CeD diagnosis. In duodenal and plasma samples, respectively, 897 and 140 DEPs were identified. Combining weighted gene co-expression network analysis(WGCNA) with the DEPs, five key proteins were identified across three machine learning methods. FGL2 and TXNDC5 were significantly elevated in the CeD group, while CHGA expression showed an increasing trend, but without statistical significance. The receiver operating characteristic curve results indicated an area under the curve (AUC) of 0.7711 for FGL2 and 0.6978 for TXNDC5, with a combined AUC of 0.8944. Exploratory analysis using Mfuzz and three machine learning methods identified four plasma proteins potentially associated with CeD pathological grading (Marsh classification): FABP, CPOX, BHMT, and PPP2CB. We conclude that FGL2 and TXNDC5 deserve exploration as potential sensitive, noninvasive diagnostic biomarkers for CeD.
Keywords: 4D-DIA proteomics; Biomarker; Celiac disease; Machine learning; Marsh classification; WGCNA.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: The project received approval from the Ethics Committee of the Xinjiang Uyghur Autonomous Region People’s Hospital (KY20220311067 and KY2023013103). Each participant was given a written informed consent form, stating that the specimens collected would be used for pathological examination and related medical research. Conflict of interest: The authors declare no conflicts of interest.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

