EPRS1-mediated fibroblast activation and mitochondrial dysfunction promote kidney fibrosis
- PMID: 39623092
- PMCID: PMC11671583
- DOI: 10.1038/s12276-024-01360-6
EPRS1-mediated fibroblast activation and mitochondrial dysfunction promote kidney fibrosis
Abstract
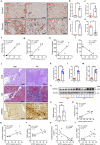
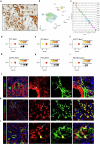
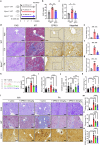
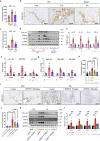
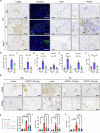
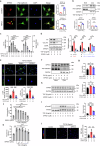
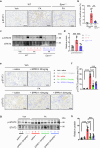
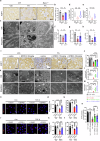
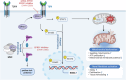
Kidney fibrosis causes irreversible structural damage in chronic kidney disease and is characterized by aberrant extracellular matrix (ECM) accumulation. Although glutamyl-prolyl-tRNA synthetase 1 (EPRS1) is a crucial enzyme involved in proline-rich protein synthesis, its role in kidney fibrosis remains unclear. The present study revealed that EPRS1 expression levels were increased in the fibrotic kidneys of patients and mice, especially in fibroblasts and proximal tubular epithelial cells, on the basis of single-cell analysis and immunostaining of fibrotic kidneys. Moreover, C57BL/6 EPRS1tm1b heterozygous knockout (Eprs1+/-) and pharmacological EPRS1 inhibition with the first-in-class EPRS1 inhibitor DWN12088 protected against kidney fibrosis and dysfunction by preventing fibroblast activation and proximal tubular injury. Interestingly, in vitro assays demonstrated that EPRS1-mediated nontranslational pathways in addition to translational pathways under transforming growth factor β-treated conditions by phosphorylating SMAD family member 3 in fibroblasts and signal transducers and activators of transcription 3 in injured proximal tubules. EPRS1 knockdown and catalytic inhibition suppressed these pathways, preventing fibroblast activation, proliferation, and subsequent collagen production. Additionally, we revealed that EPRS1 caused mitochondrial damage in proximal tubules but that this damage was attenuated by EPRS1 inhibition. Our findings suggest that the EPRS1-mediated ECM accumulation induces kidney fibrosis via fibroblast activation and mitochondrial dysfunction. Therefore, targeting EPRS1 could be a potential therapeutic target for alleviating fibrotic injury in chronic kidney disease.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
References
MeSH terms
LinkOut - more resources
Full Text Sources

