[Plasma long noncoding RNA expression profiles in patients with Parkinson's disease and the role of lnc-CTSD-5:1 in a PD cell model: a ceRNA microarray-based study]
- PMID: 39623270
- PMCID: PMC11605201
- DOI: 10.12122/j.issn.1673-4254.2024.11.11
[Plasma long noncoding RNA expression profiles in patients with Parkinson's disease and the role of lnc-CTSD-5:1 in a PD cell model: a ceRNA microarray-based study]
Abstract
Objective: To explore the key genes and long non-coding RNAs (lncRNAs) associated with Parkinson's disease (PD).
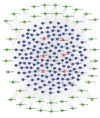
Methods: Peripheral blood plasma samples were collected from 6 PD patients and 6 healthy individuals. The mRNA and lncRNA expression profiles were detected using ceRNA microarray technology, and the differentially expressed genes were analyzed using bioinformatics methods. The differentially expressed mRNAs transcribed within 10 kb upstream or downstream of the differentially expressed lncRNAs were defined as potential cis-regulatable (Cis) target genes of the lncRNAs. A PD-specific protein-protein interaction network (PPI) was constructed. Competitive endogenous RNA (ceRNA) networks were also constructed using the differential lncRNAs with mRNAs and known microRNAs. Using MPP+-treated SHSY5Y cells as a PD cell model, the expressions of the key lncRNAs and their functions were examined.
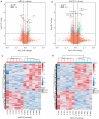
Results: We identified 316 genes and 986 lncRNAs showing significant differential expressions in PD patients (P< 0.05). The differentially expressed mRNAs and the potential cis-regulatable target genes of these lncRNAs were functionally annotated using GO and KEGG enrichment analysis, and the targeting relationship of the differentially expressed mRNAs and lncRNAs with microRNAs were predicted. Analysis of the ceRNA networks constructed based on the differentially expressed lncRNAs suggested that lncMTG2-1:1, lnc-CTSD-5:1, lnc-PCCA-3:1, lnc-VTCN1-3:1, lnc-ZNF25-7:1, and lnc-DAZ3-1:1 might be the key lncRNAs in PD. In MPP+-treated SH-SY5Y cells, the expression of lnc-CTSD-5:1 showed the most significant changes, and silencing lnc-CTSD-5:1 obviously restored the expression level of tyrosine hydroxylase.
Conclusion: PD patients have significant changes in plasma lncRNA expression profile, and the differentially expressed genes and lncRNAs found in this study may provide new clues for exploring the pathogenesis and identifying potential biomarkers of PD.
目的: 探索外周血浆中与帕金森病(PD)诊疗相关的关键基因和长链非编码RNA(lncRNA)。
方法: 收集6例PD患者和6例健康对照者的外周血血浆样本。采用竞争性内源性RNA(ceRNA)芯片技术检测mRNA和lncRNA的表达,运用生物信息学方法对差异表达的基因进行分析。将DElncRNAs上游或下游10 kb内转录的DEmRNAs定义为DElncRNAs潜在的可顺式调控的靶基因。构建PD特异性蛋白质-蛋白质相互作用网络。利用差异lncRNAs与mRNA及已知的microRNA构建ceRNA网络。采用MPP+处理SH-SY5Y细胞建立PD细胞模型,验证关键lncRNA的功能。
结果: PD患者与健康对照者间有316个基因和986个lncRNA的表达存在差异(P<0.05)。通过GO和KEGG富集分析对差异表达的mRNA和lncRNA潜在的可顺式调控的靶基因进行了功能注释,预测了差异表达的mRNA和lncRNAs与microRNA的靶向关系,并根据差异表达的lncRNA的上调或下调分别构建了ceRNA网络。基于网络分析,推测lnc-MTG2-1∶1、lnc-CTSD-5∶1、lnc-PCCA-3∶1、lnc-VTCN1-3∶1、lnc-ZNF25-7∶1和lnc-DAZ3-1∶1可能是PD的关键lncRNA。在MPP+诱导的PD细胞模型中,lnc-CTSD-5∶1的表达变化最显著(P<0.01),且其沉默有助于恢复酪氨酸羟化酶的蛋白表达水平。
结论: PD患者血浆中的lncRNA表达模式发生改变,本研究发现了新的DEGs和DElncRNAs,为探索PD的发病机制和开发潜在的生物标志物提供了新的线索。
Keywords: Parkinson's disease; bioinformatics analysis; long non-coding RNA; plasma; tyrosine hydroxylase.
Figures




References
-
- 陈芝君, 马 建, 唐 娜, 等. 中国帕金森病疾病负担变化趋势分析及预测[J]. 中国慢性病预防与控制, 2022, 30(9): 649-54.
-
- 刘永文, 焦 敏, 王 岩, 等. 1990-2019年中国帕金森病疾病负担分析[J]. 疾病监测, 2024, 39(3): 363-8.
-
- Morris HR, Spillantini MG, Sue CM, et al. . The pathogenesis of Parkinson's disease[J]. Lancet, 2024, 403(10423): 293-304. - PubMed
-
- Li K, Wang ZQ. lncRNA NEAT1: key player in neurodegenerative diseases[J]. Ageing Res Rev, 2023, 86: 101878. - PubMed
-
- Su LN, Li RQ, Zhang ZQ, et al. . Identification of altered exosomal microRNAs and mRNAs in Alzheimer's disease[J]. Ageing Res Rev, 2022, 73: 101497. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
