Expression quantitative trait loci influence DNA damage-induced apoptosis in cancer
- PMID: 39623312
- PMCID: PMC11613471
- DOI: 10.1186/s12864-024-11068-6
Expression quantitative trait loci influence DNA damage-induced apoptosis in cancer
Abstract
Background: Genomic instability and evading apoptosis are two fundamental hallmarks of cancer and closely linked to DNA damage response (DDR). By analyzing expression quantitative trait loci (eQTL) upon cell stimulation (called exposure eQTL (e2QTL)) it is possible to identify context specific gene regulatory variants and connect them to oncological diseases based on genome-wide association studies (GWAS).
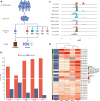
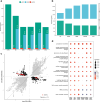
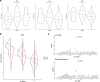
Results: We isolate CD8+ T cells from 461 healthy donors and stimulate them with high doses of 5 different carcinogens to identify regulatory mechanisms of DNA damage-induced apoptosis. Across all stimuli, we find 5,373 genes to be differentially expressed, with 85% to 99% of these genes being suppressed. While upregulated genes are specific to distinct stimuli, downregulated genes are shared across conditions but exhibit enrichment in biological processes depending on the DNA damage type. Analysis of eQTL reveals 654 regulated genes across conditions. Among them, 47 genes are significant e2QTL, representing a fraction of 4% to 5% per stimulus. To unveil disease relevant genetic variants, we compare eQTL and e2QTL with GWAS risk variants. We identify gene regulatory variants for KLF2, PIP4K2A, GPR160, RPS18, ARL17B and XBP1 that represent risk variants for oncological diseases.
Conclusion: Our study highlights the relevance of gene regulatory variants influencing DNA damage-induced apoptosis in cancer. The results provide new insights in cellular mechanisms and corresponding genes contributing to inter-individual effects in cancer development.
Keywords: Apoptosis; Cancer; DNA damage; GWAS; eQTL.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study was approved by the ethics committee of the University of Marburg and complied with the Helsinki Declaration. All participants gave written informed consent. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

