Host-specific effects of Eubacterium species on Rg3-mediated modulation of osteosarcopenia in a genetically diverse mouse population
- PMID: 39623488
- PMCID: PMC11613481
- DOI: 10.1186/s40168-024-01971-1
Host-specific effects of Eubacterium species on Rg3-mediated modulation of osteosarcopenia in a genetically diverse mouse population
Abstract
Background: Osteosarcopenia, characterized by the simultaneous loss of bone and muscle mass, is a serious health problem in the aging population. This study investigated the interplay between host genetics, gut microbiota, and musculoskeletal health in a mouse model of osteosarcopenia, exploring the therapeutic potential of gut microbiota modulation.
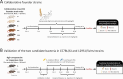
Methods: We examined the effects of Rg3, a phytochemical, on osteosarcopenia and its interactions with host genetics and gut microbiota in six founder strains of the Collaborative Cross (CC) population. Subsequently, we evaluated the therapeutic potential of Eubacterium nodatum (EN) and Eubacterium ventriosum (EV), two gut microbes identified as significant correlates of Rg3-mediated osteosarcopenia improvement, in selected C57BL/6 J (B6) and 129S1/SvImJ (129S1) mouse strains.
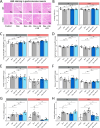
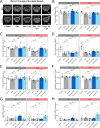
Results: Rg3 treatment altered gut microbiota composition aligned with osteosarcopenia phenotypes, which response varied depending on host genetics. This finding enabled the identification of two microbes in the Eubacterium genus, potential mediator of Rg3 effect on osteosarcopenia. Oral administration of EN and EV differentially impacted bone density, muscle mass, exercise performance, and related gene expression in a mouse strain-specific manner. In 129S1 mice, EN and EV significantly improved these parameters, effectively reversing osteosarcopenic phenotypes. Mechanistic investigations revealed that these effects were mediated through the modulation of osteoblast differentiation and protein degradation pathways. In contrast, EN and EV did not significantly improve osteosarcopenic phenotypes in B6 mice, although they did modulate mitochondrial biogenesis and microbial diversity.
Conclusions: Our findings underscore the complex interplay between host genetics and the gut microbiota in osteosarcopenia and emphasize the need for personalized treatment strategies. EN and EV exhibit strain-specific therapeutic effects, suggesting that tailoring microbial interventions to individual genetic backgrounds may be crucial for optimizing treatment outcomes. Video Abstract.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All animal experiments were approved by the International Animal Care and Use Committee of the Korea Institute of Science and Technology (Approval no.: KIST-2021–091002; Date of approval: December 30, 2021). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



References
-
- Teng Z, Zhu Y, Teng Y, Long Q, Hao Q, Yu X, et al. The analysis of osteosarcopenia as a risk factor for fractures, mortality, and falls. Osteoporos Int. 2021;32:2173–83. 10.1007/s00198-021-05963-x. - PubMed
-
- Fagundes Belchior G, Kirk B, da Pereira Silva EA, Duque G. Osteosarcopenia: beyond age-related muscle and bone loss. Eur Geriatr Med. 2020;11:715–24. 10.1007/s41999-020-00355-6. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

