Long Noncoding RNA TRIBAL Links the 8q24.13 Locus to Hepatic Lipid Metabolism and Coronary Artery Disease
- PMID: 39624902
- PMCID: PMC11651355
- DOI: 10.1161/CIRCGEN.124.004674
Long Noncoding RNA TRIBAL Links the 8q24.13 Locus to Hepatic Lipid Metabolism and Coronary Artery Disease
Abstract
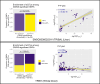
Background: Genome-wide association studies identified a 20-Kb region of chromosome 8 (8q24.13) associated with plasma lipids, hepatic steatosis, and risk for coronary artery disease. The region is proximal to TRIB1, and given its well-established role in lipid regulation in animal models, TRIB1 has been proposed to mediate the contribution of the 8q24.13 locus to these traits. This region overlaps a gene encoding the primate-specific long noncoding RNA transcript TRIBAL/TRIB1AL (TRIB1-associated locus), but the contribution of TRIBAL to coronary artery disease risk remains untested.
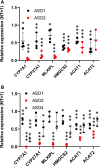
Methods: Using recently available expression quantitative trait loci data and hepatocyte models, we further investigated this locus by Mendelian randomization analysis. Following antisense oligonucleotide targeting of TRIBAL, transcription array, quantitative reverse transcription polymerase chain reaction, and enrichment analyses were performed and effects on apoB and triglyceride secretion were determined.
Results: Mendelian randomization analysis supports a causal relationship between genetically determined hepatic TRIBAL expression and markers of hepatic steatosis and coronary artery disease risk. By contrast, expression data sets did not support expression quantitative trait loci relationships between coronary artery disease-associated variants and TRIB1. TRIBAL suppression reduced the expression of key regulators of triglyceride metabolism and bile acid synthesis. Enrichment analyses identified patterns consistent with impaired metabolic functions, including reduced triglyceride and cholesterol handling ability. Furthermore, TRIBAL suppression was associated with reduced hepatocyte secretion of triglycerides.
Conclusions: This work identifies TRIBAL as a gene bridging the genotype-phenotype relationship at the 8q24.13 locus with effects on genes regulating hepatocyte lipid metabolism and triglyceride secretion.
Keywords: bile acids and salts; cholesterol; coronary artery disease; genotype; hepatocytes; lipids.
Conflict of interest statement
None.
Figures



References
-
- Chasman DI, Pare G, Mora S, Hopewell JC, Peloso G, Clarke R, Cupples LA, Hamsten A, Kathiresan S, Malarstig A, et al. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genet. 2009;5:e1000730. doi: 10.1371/journal.pgen.1000730 - PMC - PubMed
-
- Waterworth DM, Ricketts SL, Song K, Chen L, Zhao JH, Ripatti S, Aulchenko YS, Zhang W, Yuan X, Lim N, et al. ; Wellcome Trust Case Control Consortium. Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arterioscler Thromb Vasc Biol. 2010;30:2264–2276. doi: 10.1161/ATVBAHA.109.201020 - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

