Environmental fungi target thiol homeostasis to compete with Mycobacterium tuberculosis
- PMID: 39625876
- PMCID: PMC11614215
- DOI: 10.1371/journal.pbio.3002852
Environmental fungi target thiol homeostasis to compete with Mycobacterium tuberculosis
Abstract
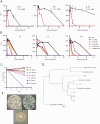
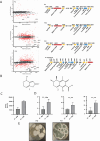
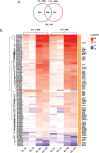
Mycobacterial species in nature are found in abundance in sphagnum peat bogs where they compete for nutrients with a variety of microorganisms including fungi. We screened a collection of fungi isolated from sphagnum bogs by co-culture with Mycobacterium tuberculosis (Mtb) to look for inducible expression of antitubercular agents and identified 5 fungi that produced cidal antitubercular agents upon exposure to live Mtb. Whole genome sequencing of these fungi followed by fungal RNAseq after Mtb exposure allowed us to identify biosynthetic gene clusters induced by co-culture. Three of these fungi induced expression of patulin, one induced citrinin expression and one induced the production of nidulalin A. The biosynthetic gene clusters for patulin and citrinin have been previously described but the genes involved in nidulalin A production have not been described before. All 3 of these potent electrophiles react with thiols and treatment of Mtb cells with these agents followed by Mtb RNAseq showed that these natural products all induce profound thiol stress suggesting a rapid depletion of mycothiol. The induction of thiol-reactive mycotoxins through 3 different systems in response to exposure to Mtb suggests that fungi have identified this as a highly vulnerable target in a similar microenvironment to that of the caseous human lesion.
Copyright: This is an open access article, free of all copyright, and may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose. The work is made available under the Creative Commons CC0 public domain dedication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Update of
-
Environmental sphagnum-associated fungi target thiol homeostasis to compete with Mycobacterium tuberculosis.bioRxiv [Preprint]. 2024 Sep 23:2024.09.23.614403. doi: 10.1101/2024.09.23.614403. bioRxiv. 2024. Update in: PLoS Biol. 2024 Dec 3;22(12):e3002852. doi: 10.1371/journal.pbio.3002852. PMID: 39372785 Free PMC article. Updated. Preprint.
References
-
- Sarathy JP, Xie M, Jones RM, Chang A, Osiecki P, Weiner D, et al.. A Novel Tool to Identify Bactericidal Compounds against Vulnerable Targets in Drug-Tolerant M. tuberculosis found in Caseum. mBio. 2023;14(2):e0059823. Epub 20230405. doi: 10.1128/mbio.00598-23 ; PubMed Central PMCID: PMC10127596. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

