Detecting global and local hierarchical structures in cell-cell communication using CrossChat
- PMID: 39627184
- PMCID: PMC11615294
- DOI: 10.1038/s41467-024-54821-x
Detecting global and local hierarchical structures in cell-cell communication using CrossChat
Abstract
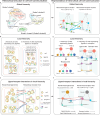
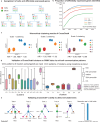
Cell-cell communication (CCC) occurs across different biological scales, ranging from interactions between large groups of cells to interactions between individual cells, forming a hierarchical structure. Globally, CCC may exist between clusters or only subgroups of a cluster with varying size, while locally, a group of cells as sender or receiver may exhibit distinct signaling properties. Current existing methods infer CCC from single-cell RNA-seq or Spatial Transcriptomics only between predefined cell groups, neglecting the existing hierarchical structure within CCC that are determined by signaling molecules, in particular, ligands and receptors. Here, we develop CrossChat, a novel computational framework designed to infer and analyze the hierarchical cell-cell communication structures using two complementary approaches: a global hierarchical structure using a multi-resolution clustering method, and multiple local hierarchical structures using a tree detection method. This framework provides a comprehensive approach to understand the hierarchical relationships within CCC that govern complex tissue functions. By applying our method to two nonspatial scRNA-seq datasets sampled from COVID-19 patients and mouse embryonic skin, and two spatial transcriptomics datasets generated from Stereo-seq of mouse embryo and 10x Visium of mouse wounded skin, we showcase CrossChat's functionalities for analyzing both global and local hierarchical structures within cell-cell communication.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
Grants and funding
- R01 DE030565/DE/NIDCR NIH HHS/United States
- P30 AR075047/AR/NIAMS NIH HHS/United States
- R01AR079150/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
- 594598/Simons Foundation
- R01DE030565/U.S. Department of Health & Human Services | NIH | National Institute of Dental and Craniofacial Research (NIDCR)
- CBET2134916/NSF | ENG/OAD | Division of Chemical, Bioengineering, Environmental, and Transport Systems (CBET)
- R01 GM152494/GM/NIGMS NIH HHS/United States
- R01 AR079150/AR/NIAMS NIH HHS/United States
- DMS1763272/NSF | Directorate for Mathematical & Physical Sciences | Division of Mathematical Sciences (DMS)
- R01GM152494/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)

