Endothelial Pim3 kinase protects the vascular barrier during lung metastasis
- PMID: 39627185
- PMCID: PMC11615401
- DOI: 10.1038/s41467-024-54445-1
Endothelial Pim3 kinase protects the vascular barrier during lung metastasis
Abstract
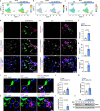
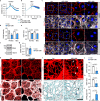
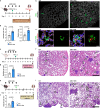
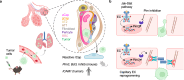
Endothelial cells (ECs) form a tissue-specific barrier for disseminating cancer cells in distant organs. However, the molecular regulation of the ECs in the metastatic niche remains unclear. Here, we analyze using scRNA-Seq, the transcriptional reprogramming of lung ECs six hours after the arrival of melanoma cells in mouse lungs. We discover a reactive capillary EC cluster (rCap) that increases from general capillary ECs in response to infiltrating cancer cells. rCap is enriched for angiogenic and inflammatory pathways and is also found in human lung datasets. The JAK-STAT activated oncogenic Pim3 kinase is a marker of rCap, being upregulated in spontaneous metastasis models. Notably, PIM inhibition increases vascular leakage and metastatic colonization and impairs the EC barrier by decreasing the junctional cadherin-5 and catenins α, β and δ. These results highlight the pulmonary endothelium's plasticity and its protection by PIM3, which may impair the efficacy of PIM inhibitors in cancer therapies.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

