Cell polarity proteins promote macropinocytosis in response to metabolic stress
- PMID: 39627191
- PMCID: PMC11614886
- DOI: 10.1038/s41467-024-54788-9
Cell polarity proteins promote macropinocytosis in response to metabolic stress
Abstract
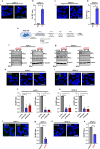
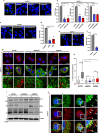
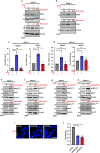
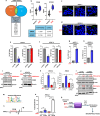
Macropinocytosis has emerged as a scavenging pathway that cancer cells exploit to survive in a nutrient-deprived microenvironment. Tumor cells are especially reliant on glutamine for their survival, and in pancreatic ductal adenocarcinoma (PDAC) cells, glutamine deficiency can enhance the stimulation of macropinocytosis. Here, we identify the atypical protein kinase C (aPKC) enzymes, PKCζ and PKCι, as regulators of macropinocytosis. In normal epithelial cells, aPKCs associate with the scaffold proteins Par3 and Par6 to regulate cell polarity, affecting several targets, including the Par1 kinases and we find that each of these proteins is required for macropinocytosis. Mechanistically, aPKCs are regulated by EGFR signaling or by the transcription factor CREM to promote the Par3 relocation to microtubules, facilitating macropinocytosis in a dynein-dependent manner. Importantly, cell fitness impairment caused by aPKC depletion is rescued by the restoration of macropinocytosis and aPKCs support PDAC growth in vivo. Our findings enhance our understanding of the mechanistic underpinnings that control macropinocytic uptake in the context of metabolic stress.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: C.C. is an inventor on a U.S. patent titled “Cancer diagnostics, therapeutics, and drug discovery associated with macropinocytosis,” Patent number: US-11209420-B2. The remaining authors declare no competing interests.
Figures
Update of
-
Cell polarity proteins promote macropinocytosis in response to metabolic stress.bioRxiv [Preprint]. 2024 Jan 30:2024.01.16.575943. doi: 10.1101/2024.01.16.575943. bioRxiv. 2024. Update in: Nat Commun. 2024 Dec 3;15(1):10541. doi: 10.1038/s41467-024-54788-9. PMID: 38293142 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

