Loss of NUMB drives aggressive bladder cancer via a RHOA/ROCK/YAP signaling axis
- PMID: 39627202
- PMCID: PMC11615365
- DOI: 10.1038/s41467-024-54246-6
Loss of NUMB drives aggressive bladder cancer via a RHOA/ROCK/YAP signaling axis
Abstract
Advances in bladder cancer (BCa) treatment have been hampered by the lack of predictive biomarkers and targeted therapies. Here, we demonstrate that loss of the tumor suppressor NUMB promotes aggressive bladder tumorigenesis and worsens disease outcomes. Retrospective cohort studies show that NUMB-loss correlates with poor prognosis in post-cystectomy muscle-invasive BCa patients and increased risk of muscle invasion progression in non-muscle invasive BCa patients. In mouse models, targeted Numb ablation induces spontaneous tumorigenesis and sensitizes the urothelium to carcinogenic insults, accelerating tumor onset and progression. Integrative transcriptomic and functional analyses in mouse and human BCa models reveal that upregulation of YAP transcriptional activity via a RHOA/ROCK-dependent pathway is a hallmark of NUMB-deficient BCa. Pharmacological or genetic inhibition of this molecular pathway selectively inhibits proliferation and invasion of NUMB-deficient BCa cells in 3D-Matrigel organoids. Thus, NUMB-loss could serve as a biomarker for identifying high-risk patients who may benefit from targeted anti-RHOA/ROCK/YAP therapies.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: G. Renne, G. Vago, D. Tosoni and S. Pece are co-inventors of a patent (EP24191940.6) related to this work, pending to the European Institute of Oncology and University of Milan. All the other authors declare no competing interests.
Figures
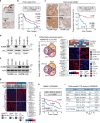
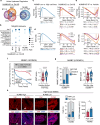

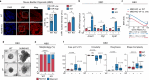


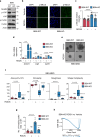

References
-
- Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin.71, 7–33 (2021). - PubMed
-
- Kaufman, D. S., Shipley, W. U. & Feldman, A. S. Bladder cancer. Lancet374, 239–249 (2009). - PubMed
-
- Kamat, A. M. et al. Bladder cancer. Lancet388, 2796–2810 (2016). - PubMed
-
- Witjes, J. A. et al. European Association of Urology Guidelines on Muscle-invasive and Metastatic Bladder Cancer: Summary of the 2020 Guidelines. Eur. Urol.79, 82–104 (2021). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- IG 23049/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- IG 23060/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- MIUR-PRIN2017/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)
- MIUR/PRIN2020/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)
- 5x1000 funds/Ministero della Salute (Ministry of Health, Italy)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

