ATAD5-BAZ1B interaction modulates PCNA ubiquitination during DNA repair
- PMID: 39627214
- PMCID: PMC11615311
- DOI: 10.1038/s41467-024-55005-3
ATAD5-BAZ1B interaction modulates PCNA ubiquitination during DNA repair
Abstract
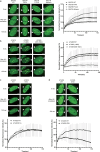
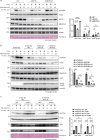
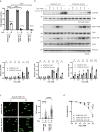
Mono-ubiquitinated PCNA (mono-Ub-PCNA) is generated when replication forks encounter obstacles, enabling the bypass of DNA lesions. After resolving stalled forks, Ub-PCNA must be de-ubiquitinated to resume high-fidelity DNA synthesis. ATAD5, in cooperation with the UAF1-USP1 complex, is responsible for this de-ubiquitination. However, the precise regulation of timely Ub-PCNA de-ubiquitination remains unclear. Our research reveals that BAZ1B, a regulatory subunit of the BAZ1B-SMARCA5 chromatin-remodeling complex (also known as the WICH complex), plays a crucial role in fine-tuning the de-ubiquitination process of Ub-PCNA. The BAZ1B binding region of ATAD5 encompasses the UAF1-binding domain of ATAD5. Disruption of the ATAD5-BAZ1B interaction results in premature de-ubiquitination of Ub-PCNA following treatment with hydrogen peroxide. Cells with impaired BAZ1B binding to ATAD5 display increased sensitivity to oxidative stress compared to wild-type cells. These findings suggest that BAZ1B prevents premature Ub-PCNA de-ubiquitination, thereby safeguarding genome integrity.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

