Spint1 disruption in mouse pancreas leads to glucose intolerance and impaired insulin production involving HEPSIN/MAFA
- PMID: 39627229
- PMCID: PMC11615295
- DOI: 10.1038/s41467-024-54927-2
Spint1 disruption in mouse pancreas leads to glucose intolerance and impaired insulin production involving HEPSIN/MAFA
Abstract
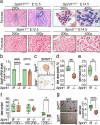
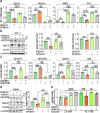
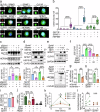
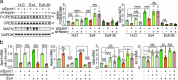
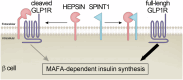
SPINT1, a membrane-anchored serine protease inhibitor, regulates cascades of pericellular proteolysis while its tissue-specific functions remain incompletely characterized. In this study, we generate Spint1-lacZ knock-in mice and observe Spint1 expression in embryonic pancreatic epithelium. Pancreas-specific Spint1 disruption significantly diminishes islet size and mass, causing glucose intolerance and downregulation of MAFA and insulin. Mechanistically, the serine protease HEPSIN interacts with SPINT1 in β cells, and Hepsin silencing counteracts the downregulation of Mafa and Ins1 caused by Spint1 depletion. Furthermore, we demonstrate a potential interaction between HEPSIN and GLP1R in β cells. Spint1 silencing or Hepsin overexpression reduces GLP1R-related cyclic AMP levels and Mafa expression. Spint1-disrupted mice also exhibit a significant reduction in Exendin-4-induced insulin secretion. Moreover, SPINT1 expression increases in islets of prediabetic humans compared to non-prediabetic groups. The results unveil a role for SPINT1 in β cells, modulating glucose homeostasis and insulin production via HEPSIN/MAFA signaling.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Huang, H.-P. et al. Persistent elevation of hepatocyte growth factor activator inhibitors in cholangiopathies affects liver fibrosis and differentiation. Hepatology55, 161–72 (2012). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

