SARS-CoV-2 Nsp6-Omicron causes less damage to the Drosophila heart and mouse cardiomyocytes than ancestral Nsp6
- PMID: 39627475
- PMCID: PMC11615247
- DOI: 10.1038/s42003-024-07307-x
SARS-CoV-2 Nsp6-Omicron causes less damage to the Drosophila heart and mouse cardiomyocytes than ancestral Nsp6
Abstract
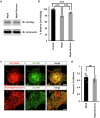
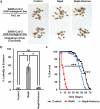
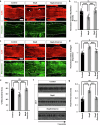
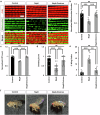
A few years into the COVID-19 pandemic, the SARS-CoV-2 Omicron strain rapidly becomes and has remained the predominant strain. To date, Omicron and its subvariants, while more transmittable, appear to cause less severe disease than prior strains. To study the cause of this reduced pathogenicity we compare SARS-CoV-2 ancestral Nsp6 with Nsp6-Omicron, which we have previously identified as one of the most pathogenic viral proteins. Here, through ubiquitous expression in Drosophila, we show that ancestral Nsp6 causes both structural and functional damage to cardiac, muscular, and tracheal (lung) tissue, whereas Nsp6-Omicron has minimal effects. Moreover, we show that ancestral Nsp6 dysregulates the glycolysis pathway and disrupts mitochondrial function, whereas Nsp6-Omicron does not. Through validation in mouse primary cardiomyocytes, we find that Nsp6-induced dysregulated glycolysis underlies the cardiac dysfunction. Together, the results indicate that the amino acid changes in Omicron might hinder its interaction with host proteins thereby minimizing its pathogenicity.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
SARS-CoV-2 Nsp6 damages Drosophila heart and mouse cardiomyocytes through MGA/MAX complex-mediated increased glycolysis.Commun Biol. 2022 Sep 30;5(1):1039. doi: 10.1038/s42003-022-03986-6. Commun Biol. 2022. PMID: 36180527 Free PMC article.
-
SARS-CoV-2 nonstructural protein 6 from Alpha to Omicron: evolution of a transmembrane protein.mBio. 2023 Aug 31;14(4):e0068823. doi: 10.1128/mbio.00688-23. Epub 2023 Jul 21. mBio. 2023. PMID: 37477426 Free PMC article.
-
NSP6 of SARS-CoV-2 Dually Regulates Autophagic-Lysosomal Degradation.Int J Mol Sci. 2025 Apr 14;26(8):3699. doi: 10.3390/ijms26083699. Int J Mol Sci. 2025. PMID: 40332329 Free PMC article.
-
Unique mutations in SARS-CoV-2 Omicron subvariants' non-spike proteins: Potential impacts on viral pathogenesis and host immune evasion.Microb Pathog. 2022 Sep;170:105699. doi: 10.1016/j.micpath.2022.105699. Epub 2022 Aug 6. Microb Pathog. 2022. PMID: 35944840 Free PMC article. Review.
-
Omicron: What Makes the Latest SARS-CoV-2 Variant of Concern So Concerning?J Virol. 2022 Mar 23;96(6):e0207721. doi: 10.1128/jvi.02077-21. Epub 2022 Mar 23. J Virol. 2022. PMID: 35225672 Free PMC article. Review.
References
-
- Centers for Disease Control and Prevention. COVID Data Tracker. CDC,https://covid.cdc.gov/covid-data-tracker/#variant-summary (2024).
-
- European Centre for Disease Prevention and Control-An agency of the European Union. SARS-CoV-2 variants of concern as of 16 February 2024. ECDC,https://www.ecdc.europa.eu/en/covid-19/variants-concern (2024).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

