Unique structural configuration of EV-DNA primes Kupffer cell-mediated antitumor immunity to prevent metastatic progression
- PMID: 39627554
- PMCID: PMC12185067
- DOI: 10.1038/s43018-024-00862-6
Unique structural configuration of EV-DNA primes Kupffer cell-mediated antitumor immunity to prevent metastatic progression
Abstract
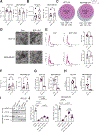
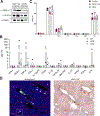
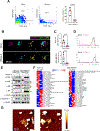
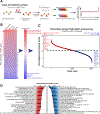
Extracellular vesicles (EVs) transport biomolecules that mediate intercellular communication. We previously showed that EVs contain DNA (EV-DNA) representing the entire genome. However, the mechanism of genomic EV-DNA packaging and its role in cancer remain elusive. We now demonstrate that EV-DNA is predominantly localized on the vesicle surface and associated with uniquely modified and cleaved histones. Moreover, a genome-wide clustered regularly interspaced short palindromic repeats knockout screen revealed that immune developmental pathways and genes, including apoptotic peptidase activating factor 1 (APAF1) and neutrophil cytosolic factor 1 (NCF1), regulate EV-DNA packaging. Furthermore, in colorectal cancer models, uptake of EV-DNA by pre-metastatic liver Kupffer cells (KCs) activated DNA damage responses. This activation rewired KC cytokine production and promoted the formation of tertiary lymphoid structures, thereby suppressing liver metastasis. Conversely, loss of APAF1 decreased EV-DNA packaging and promoted liver metastasis. Importantly, colorectal cancer biopsy EV-DNA secretion could serve as a predictive biomarker for postoperative metastasis. Taken together, our findings indicate that uniquely chromatinized EV-DNA induces antitumor immunity.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests: D.L. is on the scientific advisory board of Aufbau Holdings. R.E.S. is on the scientific advisory board of Miromatrix and is a speaker and consultant for Alnylam. The remaining authors declare no competing interests.
Figures






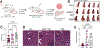

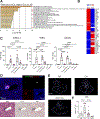

References
MeSH terms
Substances
Grants and funding
- 23-0105/AICR_/Worldwide Cancer Research/United Kingdom
- 20012443/Ministry of Trade, Industry and Energy (Ministry of Trade, Industry and Energy, Korea)
- P50 CA192937/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- HI19C1330/Korea Health Industry Development Institute (KHIDI)
- U01 CA210240/CA/NCI NIH HHS/United States
- CA218513/Center for Strategic Scientific Initiatives, National Cancer Institute (NCI Center for Strategic Scientific Initiatives)
- R35 CA232093/CA/NCI NIH HHS/United States
- CA224175 (D.L.), CA210240 (D.L.), CA232093 (D.L.), CA207983 (D.L.), CA169538 (D.L.) and CA218513/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01 CA218513/CA/NCI NIH HHS/United States
- R01 CA207983/CA/NCI NIH HHS/United States
- R35 GM138386/GM/NIGMS NIH HHS/United States
- U01 CA169538/CA/NCI NIH HHS/United States
- U01 CA224175/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

