Receptor tyrosine kinase inhibition leads to regression of acral melanoma by targeting the tumor microenvironment
- PMID: 39627834
- PMCID: PMC11613472
- DOI: 10.1186/s13046-024-03234-1
Receptor tyrosine kinase inhibition leads to regression of acral melanoma by targeting the tumor microenvironment
Abstract
Background: Acral melanoma (AM) is an aggressive melanoma variant that arises from palmar, plantar, and nail unit melanocytes. Compared to non-acral cutaneous melanoma (CM), AM is biologically distinct, has an equal incidence across genetic ancestries, typically presents in advanced stage disease, is less responsive to therapy, and has an overall worse prognosis.
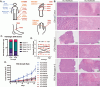
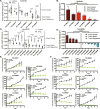
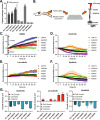
Methods: An independent analysis of published sequencing data was performed to evaluate the frequency of receptor tyrosine kinase (RTK) ligands and adapter protein gene variants and expression. To target these genetic variants, a zebrafish acral melanoma model and preclinical patient-derived xenograft (PDX) mouse models were treated with a panel of RTK inhibitors. Residual PDX tumors were evaluated for changes in proliferation, vasculature, necrosis, and ferroptosis by histology and immunohistochemistry.
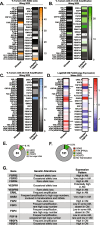
Results: RTK ligands and adapter proteins are frequently amplified, translocated, and/or overexpressed in AM. Dual FGFR/VEGFR inhibitors decrease acral-analogous melanocyte proliferation and migration in zebrafish, and the potent pan-FGFR/VEGFR inhibitor, Lenvatinib, uniformly induces tumor regression in AM PDX tumors but only slows tumor growth in CM models. Unlike other multi-RTK inhibitors, Lenvatinib is not directly cytotoxic to dissociated AM PDX tumor cells and instead disrupts tumor architecture and vascular networks.
Conclusion: Considering the great difficulty in establishing AM cell culture lines, these findings suggest that AM may be more sensitive to microenvironment perturbations than CM. In conclusion, dual FGFR/VEGFR inhibition may be a viable therapeutic strategy that targets the unique biology of AM.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Tumor tissue was obtained from patients who provided written informed consent according to a tissue collection protocol (University of Utah IRB 89989 and 10924) approved by the Huntsman Cancer. Institute (HCI) Institutional Review Board. Clinical chart review, slide procurement, and slide imaging were performed under the ARUP umbrella IRB protocol (#00091019) for general pathology specimens and the Dermatopathology umbrella IRB protocol (#00076927) for dermatopathology specimens. Consent for publication: All authors have consented to the publication of this article. Competing interests: The authors declare no conflict of interest.
Figures


Update of
-
Receptor tyrosine kinase inhibition leads to regression of acral melanoma by targeting the tumor microenvironment.bioRxiv [Preprint]. 2024 Jun 17:2024.06.15.599116. doi: 10.1101/2024.06.15.599116. bioRxiv. 2024. Update in: J Exp Clin Cancer Res. 2024 Dec 3;43(1):317. doi: 10.1186/s13046-024-03234-1. PMID: 38948879 Free PMC article. Updated. Preprint.
References
-
- Darmawan CC, Jo G, Montenegro SE, Kwak Y, Cheol L, Cho KH, et al. Early detection of acral melanoma: A review of clinical, dermoscopic, histopathologic, and molecular characteristics. J Am Acad Dermatol. 2019:805–12. 10.1016/j.jaad.2019.01.081. - PubMed
-
- Basurto-Lozada P, Molina-Aguilar C, Castaneda-Garcia C, Vázquez-Cruz ME, Garcia-Salinas OI, Álvarez-Cano A, et al. Acral lentiginous melanoma: Basic facts, biological characteristics and research perspectives of an understudied disease. Pigment Cell Melanoma Res. 2021;34:59–71. 10.1111/pcmr.12885. - PMC - PubMed
-
- Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545:175–80. 10.1038/nature22071. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

